The immobilization of proteins on a supporting substrate is necessary for many important research endeavors. The construction of biosensors for diagnostics, for the observation and development of enzymatic processes, or for sensitive drug screening requires the immobilization of specific proteins such as antibodies and receptors, and protein arrays are needed for studies in the field of proteomics.
Among the considerations important when preparing an immobilized protein array is the cost, which of course increases with the number of labor-intensive steps. Anyone who has worked in a laboratory also knows that the expense of an experiment rises alongside the extent of purification of the samples or chemicals being used. Selectivity – the ability to immobilize only specific proteins onto the substrate – is also critical for achieving a good sensitivity of the array to a specific target without producing false positive or negative results, and this is not so readily achieved without purification.
Or is it?
A team at the Gwangju Institute of Science and Technology, Korea, has developed a new process for protein immobilization directly from cell lysates that avoids all the expensive purification steps but maintains target selectivity.
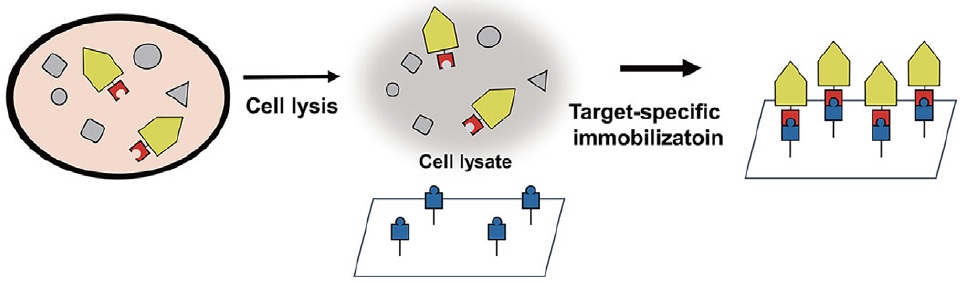
The p-azidophenylalanine (red) only incorporated into the target proteins (yellow), allowing target-specific protein immobilization in the following steps.
The team functionalized their supporting substrates with alkynes, then incorporated the clickable, non-natural amino acid p-azidophenylalanine into the target proteins and used strain-promoted alkyne–azide cycloaddition to encourage their attachment to the substrate. The p-azidophenylalanine contains a bio-orthogonal functional group, which allowed the researchers to freely choose a desired immobilization site on the protein rather than being limited to a particular N- or C-terminal region, as has been the case in previous attempts. This also means that the protein’s activity can be well maintained after immobilization – another critical consideration for practical applications.
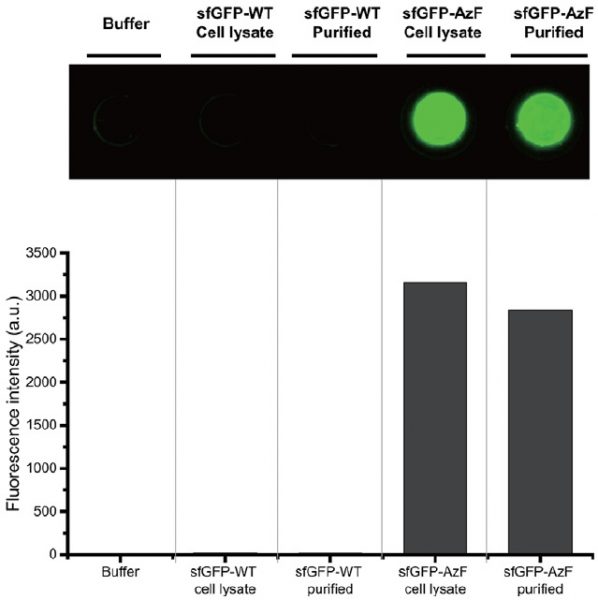
Green fluorescent protein both with (sfGFP-AzF) and without (sfGFP-WT) the p-azidophenylalanine were compared. The selectivity of the new immobilization process to p-azidophenylalanine-tagged proteins is shown to be as effective as the more expensive protein purification method.
Non-natural amino acids are generally incorporated at sites terminated with amber codons, of which there are typically plenty in the E. Coli cells used as the hosts in this process. To achieve specificity, the researchers engineered an E. Coli strain without any amber codons, meaning that the p-azidophenylalanine was not able to incorporate into anything but the exogenous target proteins, avoiding nonspecific attachment in the following steps by endogenous non-target proteins.
“Immobilization efficiency of the target protein from cell lysates was comparable to that of the purified target protein,” claim the researchers. Their technique is also generalizable. To try applying it to your research, see the details here in their Research Article published in Biotechnology Journal.
Interested in protein research? Doing some yourself? These articles may be useful to you: