Presently, there are vigorous activities in the engineering of nanomaterials for biomedical applications, e.g., as biosensors, cancer therapeutics or drug carriers. It is quite obvious that safe use of such nanomaterials must be based on a profound understanding, on the molecular level, of their interplay with biomolecules in the human body. Surfaces of engineered nanoparticles (NPs) are generally reactive toward biomolecules and complete suppression of these interactions is extremely difficult, if not impossible.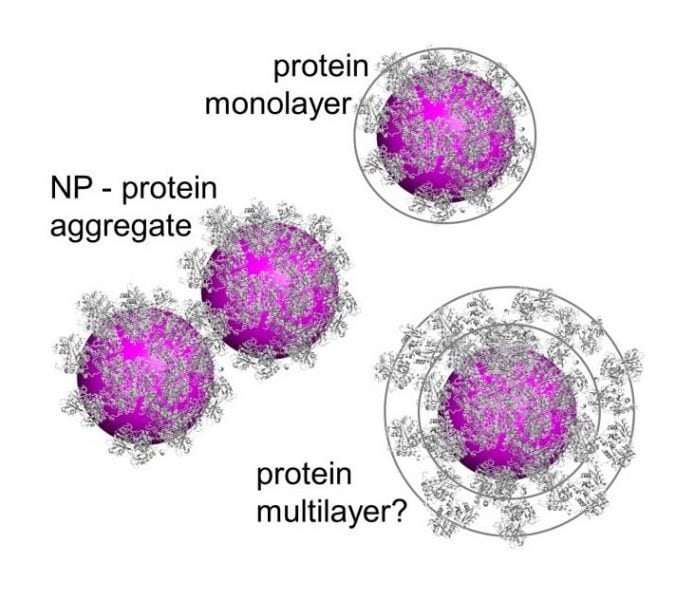
Proteins and other biomolecules bind to the NP surface, forming an adsorption layer (biomolecular corona) that modifies the NPs’ physicochemical properties and defines their interactions with living systems. In spite of significant advances in our understanding of the biomolecular corona in recent years, there are still divergent views of its microscopic structure and dynamics. In a recent Opinion piece in WIREs Nanomedicine and Nanobiotechnology, Nienhaus and co-workers focus on measurements of the corona thickness and the key question of whether proteins form monolayers or multilayers upon adsorption onto NPs.
Exploration of the biomolecular corona is a formidable endeavor, owing to the enormous diversity of engineered NPs in terms of their physicochemical properties and the vast number of biomolecules present in biofluids that may bind to NPs with widely different strengths. Depending on the interaction strengths, coronae can be dynamic (reversible) or static (irreversible) on a chosen experimental time scale. Static coronas are simpler to handle because they can be analyzed after separation of NPs and immersion fluid. Dynamic coronas; however, require in situ studies of NPs in the immersion fluid (e.g., protein solutions or blood), for which only a few, somewhat challenging methods exist.
Nienhaus and coworkers critically discuss in situ biomolecular corona characterization by means of diffusion-based nanoparticle sizing experiments, fluorescence correlation spectroscopy (FCS) and dynamic light scattering (DLS), both of which directly determine the hydrodynamic radius of NPs in a fluid via the Stokes–Einstein relation. However, these techniques are not straightforward to use and may lead non-experts to erroneous conclusions. Their size resolution is poor, and problems arise in experiments on heterogeneous samples, i.e., if the NPs tend to form small or larger aggregates.
In quantitative FCS studies of NP-protein interactions with highly defined model NPs and important serum proteins as well as blood serum, Nienhaus and co-workers so far universally found evidence of protein monolayer formation around NPs under saturation conditions. In this work, they show that formation of small NP clusters in the solution may give rise to FCS or DLS data that can be misinterpreted to indicate a thick protein corona. Therefore, straightforward routine application of these methods only works for ideal samples, and great care has to be exercised when dealing with real NP preparations. Careful, quantitative studies with a wide range of complementary techniques will produce results with a high confidence level, which are required to thoroughly elucidate the mechanisms of biomolecular corona formation and its structure.
Kindly contributed by the Authors.