According to the central dogma of molecular biology, DNA is transcribed into mRNAs and these are translated into proteins. This fundamental process is precisely orchestrated in each eukaryotic cell to determine where, when and how much protein is synthesized. This holds true for both single-cell organisms like fungal microorganisms and multicellular organisms like plants and animals.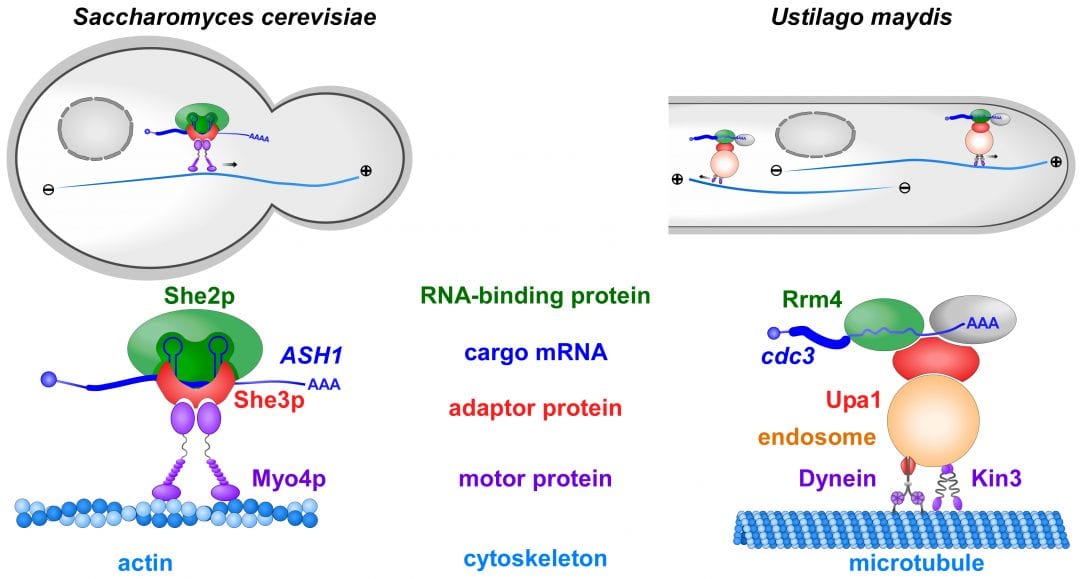
In order to determine the correct spatio-temporal expression, mRNAs are actively transported within the cell to defined subcellular sites. Comparable to postal (or zip) codes that allow the precise guidance of letters to their addressees, mRNAs contain RNA sequences that are recognizable by cellular distributors called RNA-binding proteins. RNA-binding proteins are connected to molecular motors transporting mRNAs along defined cellular tracks, the actin or microtubule cytoskeleton.
In a recent review article in WIREs RNA, Niessing and colleagues summarize the latest advancements in mRNA transport. They focus on well-studied fungal model systems like the baker’s yeast Saccharomyces cerevisiae and the plant pathogen Ustilago maydis. Highlights include the novel finding that more than one cellular distributor is needed to specifically recognize the correct postal codes in cargo mRNA.
Furthermore, an unexpected co-transport of mRNAs and membrane-enclosed organelles was recently uncovered. This not only reveals a new level of complexity but also constitutes an important step towards understanding intracellular trafficking beyond mRNA transport. Fungal organisms hence serve as “top models” for plants and animals that provide groundbreaking knowledge on these evolutionarily conserved transport processes.
Kindly contributed by the Authors.