“Is there no way to make the electron microscope more powerful?” was one of the questions that Richard Feynman asked in his famous 1959 talk “There’s plenty of room at the bottom”. Prof. Feynman claimed, among other things, that the electron microscopes of the day were “one hundred times too poor,” and that their performance was far from being limited by electron diffraction.
Since then, electron microscopy societies have flourished, and campaigns have been undertaken to either to conquer lens aberrations or to suppress the wavelength of electrons in the microscope. Both, however, require rapid advancement of expensive supporting hardware components of existing electron microscopes. But things can be dramatically different, should you think in a smarter way. Prof. John Cowley later proposed that, when an electron beam passes through a single heavy atom or a row of atoms parallel to the beam (an “atomic focuser”), a very fine beam of diameter less than 0.05 nm, is formed in the region within 1 or 2 nm of the atomic focuser. This very fine beam from the near-field may be used in scanning transmission electron microscopy or in conventional transmission electron microscopy to give image resolutions of the order of 0.05 nm, while requiring relatively minor modifications of existing instruments.
Bright ideas often rewrite a brighter history.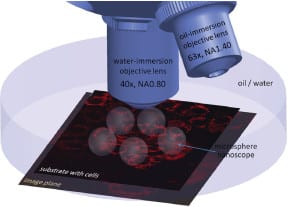
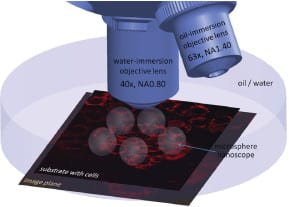
Now, researchers at the École Polytechnique Fédérale de Lausanne have successfully demonstrated super-resolution biological microscopy using virtual imaging with a simple and affordable microsphere nanoscope. The schematic, showing several microsphere nanoscopes combined with an immersion objective, illustrates that the transparent glass microspheres are located on top of cells and project near-field optical information into the far-field. This generates magnified virtual images that are observable by a classical microscope objective – which can resolve sub-diffraction features – and acts as a superlens in water or oil.
In their fluorescence experiments performed in water, the group showed that an object with a size as small as ∼λ/7, i.e. one fourth of the diffraction limit, can be quantitatively determined, while a 5.4 × magnification of the image is obtained. In addition, the potential of the technique is revealed by resolving the structure of fluorescently stained centrioles, mitochondria, chromosomes, and study the effect of doxycycline treatment on mitochondrial encoded protein expression in a mouse liver cell line. Finally, the group leader Professor Martin Gijs and his co-workers are optimistic that in future, “due to the straightforwardness of this approach, the microsphere nanoscope will be a robust and versatile tool that can be used to image with a conventional microscope a variety of biological objects such as viruses, nucleic acids, and functional organelles in living cells, and this at virtually no extra cost.”