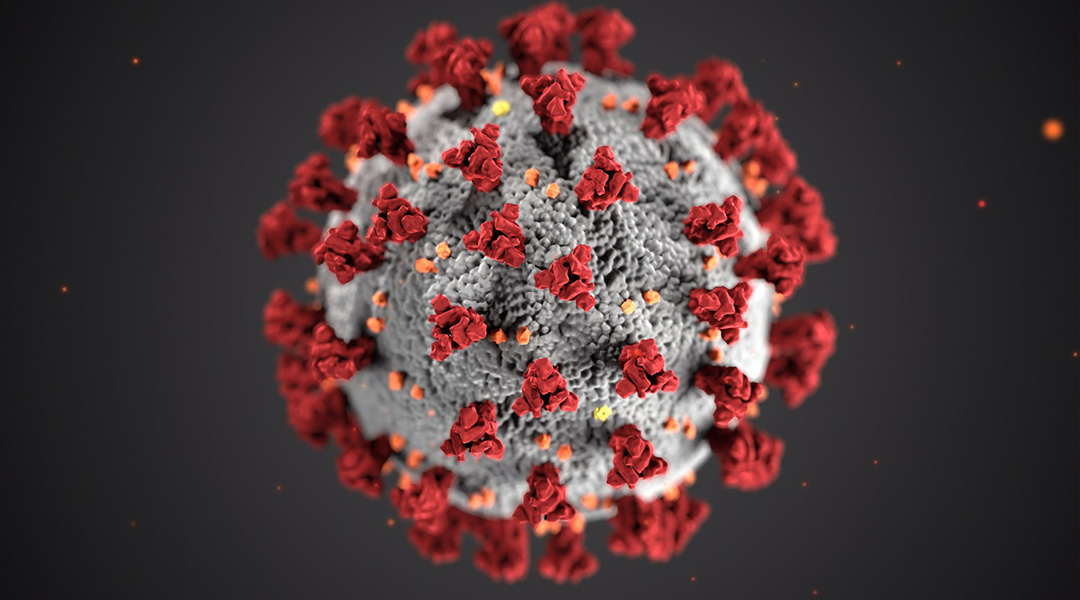
Researchers at the University of Texas at Austin and the Texas Advanced Computing Center (TACC) have taken the first steps in simulating an all-atom model of the outer envelope of the SARS-CoV-2 virus. This model is expected to give insight into how the virus infects the body right down to the molecular and atomic level.
With an estimated 200 million atoms in this model — all of whose interatomic interactions need to be calculated — this is an exceptionally daunting task. But the project’s lead, Dr. Rommie Amaro of UC San Diego, has the help of Frontera, one of the world’s most powerful supercomputers housed at TACC.
She is also not new to such feats, having done an all-atom simulation of the envelope of the influenza virus earlier this year in a paper published in ACS Central Science, which helped to identify a potentially new substrate binding mechanism.
Speaking in a press release by TACC, Amaro explains how and why these models can be important. “These simulations will give us new insights into the different parts of the coronavirus that are required for infectivity. And why we care about that is because if we can understand these different features, scientists have a better chance to design new drugs; to understand how current drugs work and potential drug combinations work.”
This is particularly important as researchers around the world scramble to trial current antiviral drugs to help treat COVID-19 patients.
Her team used molecular dynamics simulations using the NAMD code to first model individual components of the coronavirus, and ensure that each of these components are stable in the model. “Then we can introduce them into the bigger envelope simulations with neighboring molecules,” Amaro said.
Amaro is proud at how fast her team have got their COVID-19 model off the ground. It took more than a year for their influenza model to be built and run on a supercomputer. In contrast, the COVID-19 model has taken only a number of weeks, in part helped by their experience with the influenza model. “For influenza, we used the Blue Waters supercomputer, which was in some ways the predecessor to Frontera. The work, however, with the coronavirus obviously is proceeding at a much, much faster pace. This is enabled, in part because of the work that we did on Blue Waters earlier.”
This article was corrected on March 26, 2020, to correct Dr. Amaro’s affiliation to UC San Diego.