Macrophages, originally described as phagocytes, are now found to have multiple roles in innate immunity, inflammation, lipid metabolism, tissue homeostasis, injury repair, and regeneration. The functional impairment of macrophages may lead to infection, autoimmune diseases, cancer, obesity, diabetes, and cardiovascular diseases.
Human peripheral blood monocyte-derived macrophages (HMDMs) and macrophages derived from cancer cell lines, such as THP-1 and U937, serve as the main experimental models to study human macrophage biology, but HMDMs are not renewable and cancer cell lines are karyotypically abnormal. The breakthrough of induced pluripotent stem cell (iPSC) technology and their differentiation into specific cell types, including macrophages, offer a scalable experimental model to study macrophage biology and related diseases.
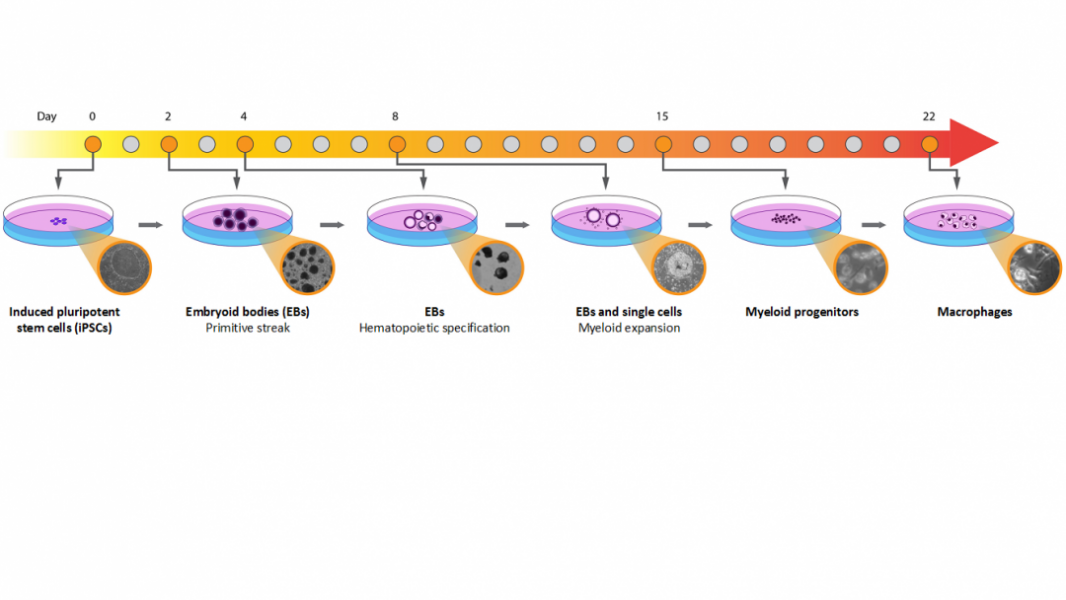
The laboratory of Dr. Hanrui Zhang at Columbia University Medical Center has developed a method to differentiate human iPSCs into macrophages, published in Current Protocols in Stem Cell Biology. It uses an embryoid body (EB)-based approach with a combination of serum-free conditions for efficient hematopoiesis specification, followed by adherent culture with serum and recombinant human macrophage colony stimulating factor (M-CSF) for myeloid expansion and macrophage maturation with high yield.
iPSC-derived macrophages (IPSDM) obtained using this protocol have demonstrated excellent functional and transcriptomic fidelity relative to human HMDM, with advantages and successful applications in disease modeling using patient-derived and CRISPR-edited iPSC lines.
Through deep RNA-sequencing, the coding transcriptome, alternative splicing events, and long non-coding RNA profiles between isogenic IPSDM and HMDM at baseline and during activation are characterized, providing a comprehensive resource for planning IPSDM studies to model such events.
This protocol and the associated resources provide a unique platform to understand human macrophage-specific functional, transcriptomic, and genomic features in physiology and a broad spectrum of macrophage-related diseases.