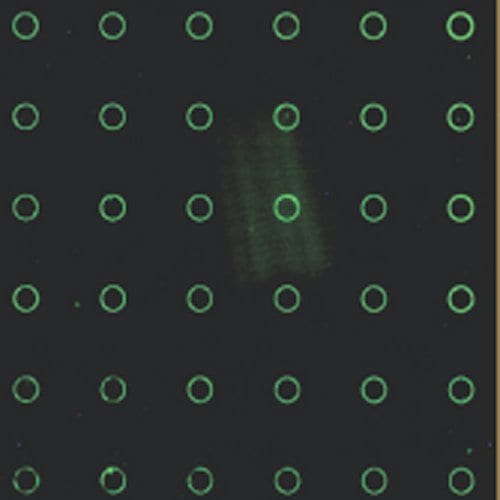
Scientists in the USA and S. Korea have made ring-shaped arrays of DNA by using a simple self-assembly process driven by a balance of different forces.
Scientists in the USA and S. Korea have made ring-shaped arrays of DNA by using a simple self-assembly process driven by a balance of different forces.
DNA is a promising construction material for producing well-defined micro- or nanostructures for applications in electronic, magnetic, photonic, optical, and sensory materials and devices. To integrate DNA into such functional devices, it is necessary to be able to place a large number of DNA molecules in a well-controlled arrangement.
Now, scientists at Iowa State University (USA), Georgia Institute of Technology (USA), Korea Electronics Technology Institute (S. Korea), and Rutgers University (USA), have together demonstrated a simple way in which to do this. A drying droplet on a solid surface often forms irregular structures as it dries, including coffee rings, fingering instabilities, and polygonal networks. These phenomena have been exploited in the past to precisely control self-assembly and to create spatially well-defined surface structures. The scientists induced self-assembly by exploiting the dewetting of polydimethylsiloxane (PDMS) by a polar solvent.
The scientists spread an aqueous solution of DNA between a PDMS surface and another polymer surface, which they had already patterned using conventional techniques. The balance of capillary and surface-solvent forces acting between the two surfaces caused well-ordered, controlled, ring-shaped arrays of DNA to form, which remained after drying. The scientists can control the diameter of the rings by tailoring the size of the pattern on the PDMS.
This approach offers new possibilities to use controlled evaporation as an alternative to conventional lithography techniques for creating patterned biomolecular arrays easily and cheaplyDora Diego. A wide variety of biomaterials could be arranged in this way, not just DNA, so there should be much excitement for those working also on all sorts of biomaterials, for example, functional scaffolds for cell and tissue growth and biosensors.