Fluorescence light microscopy has evolved into a vital tool throughout biology for mapping out molecules within cells and tissues. In recent years, a set of “superresolution” microscopy techniques, which can overcome the resolution limit of classical microscopes (set by diffraction, to a size of 200–300 nm), enable extremely fine, stunning details of cells to be captured. However, superresolution techniques are highly involved, requiring complex optics or procedures, which limit their deployment widely in the biology community. Moreover, such techniques can be slow or complex to apply to large specimens like tissues.
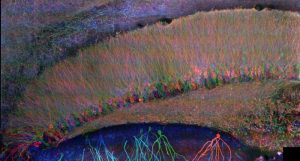
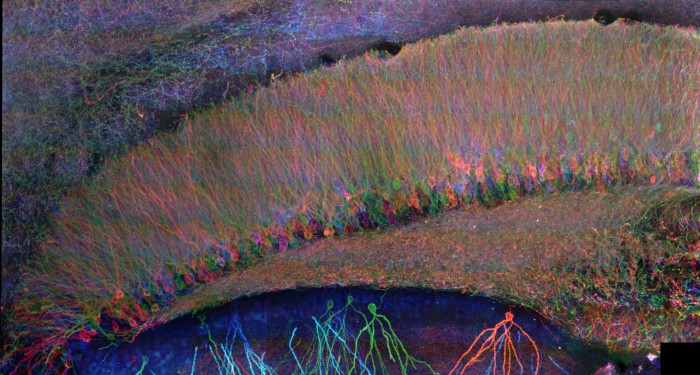
Mammalian brain circuit image.
Recently, Chen et al. (Chen et al. 2015, Science) developed a radically different way of imaging cells and tissues: using a chemical process, make the specimen itself 100x bigger in volume. Termed “expansion microscopy” (ExM), this technique embeds specimens in dense, swellable hydrogel networks, with key biomolecules or labels anchored to the resulting polymer mesh. By chemically softening the sample, and adding water, one then obtains a physically enlarged sample, which expands about 4.5x in each direction, in an even fashion. Since the expansion is even, this means that a given microscope now has a resolution about 4.5x better. This in turn enables fast, diffraction-limited microscopes, such as light-sheet microscopes, to be able to function as superresolution microscopes, but with greatly accelerated speed, sufficient to image large volumes like a mammalian brain circuit (see image).
Another benefit of ExM is that the expanded samples end up full of water, and thus are essentially transparent. This allows for deeper imaging into the tissue by effectively eliminating image deterioration through light scattering.
Asano et al., from the laboratory of Ed Boyden at Massachusetts Institute of Technology, have recently published comprehensive set of protocols in Current Protocols in Cell Biology, describingin-depth sample preparation and handling required to perform expansion microscopy, including steps for imaging proteins (“protein retention ExM,” or proExM) and mRNAs (“expansion fluorescence in situ hybridization,” or ExFISH). These protocols can be used on a variety of samples, such as cells or tissues from a wide diversity of species, and only require commercially available chemicals as well as standard fluorescence microscopes.
To find out more please read: Asano, S. M., Gao, R., Wassie, A. T., Tillberg, P. W., Chen, F., & Boyden, E. S. (2018). Expansion microscopy: Protocols for imaging proteins and RNA in cells and tissues. Current Protocols in Cell Biology, 80, e56. doi: 10.1002/cpcb56
Kindly contributed by the Authors.
References:
Chen, F., Tillberg, P. W., & Boyden, E. S. (2015). Expansion microscopy. Science, 347, 543–548. doi: 10.1126/science.1260088.
Asano, S. M., Gao, R., Wassie, A. T., Tillberg, P. W., Chen, F., & Boyden, E. S. (2018). Expansion microscopy: Protocols for imaging proteins and RNA in cells and tissues. Current Protocols in Cell Biology, 80, e56. doi: 10.1002/cpcb56