The actions and responses of all living organisms are influenced both by external cues and by internal states, and the same is true of cells. Cells often use large transmembrane proteins to sense and react to their environment; for example, the receptor tyrosine kinases (RTK), of which twenty families exist. These receptors are essential to development and organ growth, and their dysregulation is linked to diseases such as cancer and developmental disorders. Because RTKs are embedded within the cell membrane, one end is outside the cell to sense the surrounding environment (the “receptor” side), while the other end is inside the cell and transmits signals into the cell upon activation of the receptor (the “tyrosine kinase” side). In this way, they act as the interface between the cell’s immediate surroundings (the microenvironment) and internal regulatory networks (the cell state), playing a pivotal role in the cell’s response to stimuli and ultimate changes in behavior.
The external (microenvironmental) and internal (cell state) factors that regulate RTKs are collectively known as the “context,” and the context can change both the sensing of the signal and the response induced by the signal. This context includes: extracellular protein messengers (ligands) that bind to their respective RTKs; the receptors themselves and co-receptors that assist in RTK activation; transport of the receptors to various locations within the cell (trafficking); and transmission of signals inside the cell through downstream signaling networks that ultimately cause changes in the cell’s fate. Because there are so many contextual components that can impact RTK networks, systems biology techniques (that can measure these many components) and network-based approaches (that can map these components and their interactions) are essential to understanding—and ultimately, harnessing for therapeutic purposes—this complexity.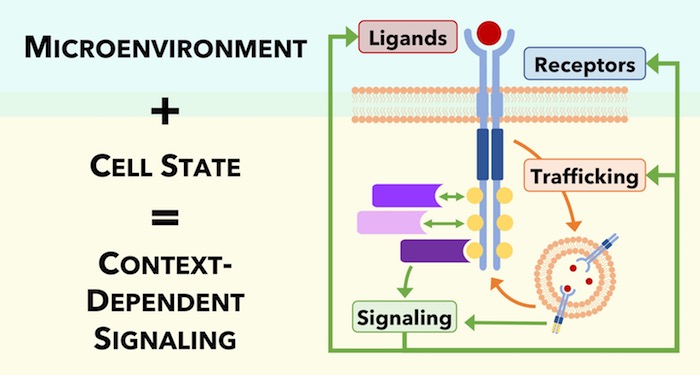
In a WIREs Systems Biology and Medicine Review, Inez Lam, Christina Pickering, and Feilim Mac Gabhann describe these different contextual components through the lens of systems biology, in which both computational methods (such as mechanistic modeling) and experimental techniques (such as high-throughput, “omics” experiments) have been used to better understand RTK systems. In particular, we describe in detail three concrete examples, involving different RTK families, that illustrate how systems approaches can elucidate the underlying biological mechanisms and reveal key insights into potential therapies targeting RTK networks.
Kindly contributed by the Authors.