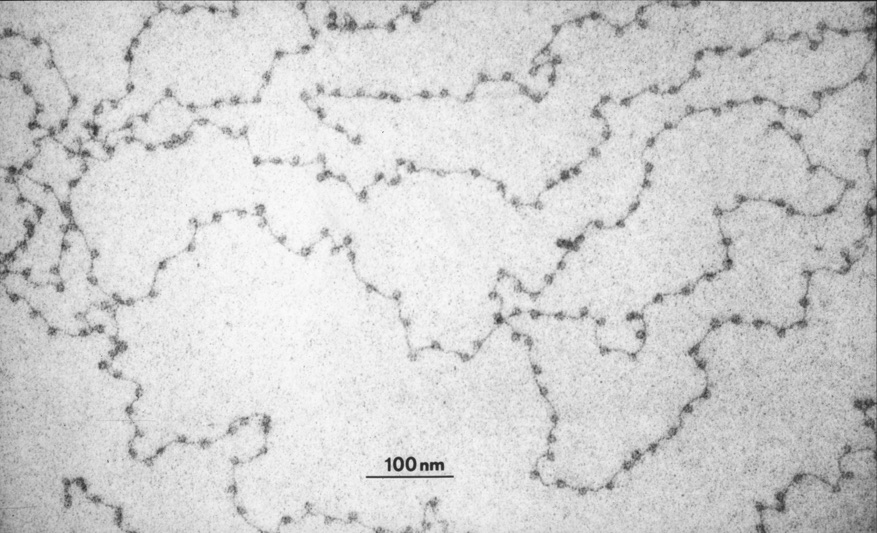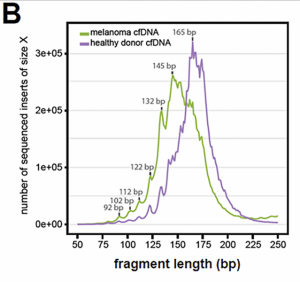
Fig 3B (from Ref3) Cancers have more diverse cell-free DNA fragments than normal cells. From this figure t is clear that the cell-free DNA fragments have a 10bp ladder-like distribution. Sequences from this individual with a melanoma are generally shorter than those from the unaffected individual. (Reproduced under open CC-BY license).
It is now possible to distinguish the pattern of DNA fragments of normal cells from cancerous cells within the blood of individuals with surgically-operable early stage cancers.1 Regions of the genome have their own local differences in cancer-specific, position-dependent patterns of fragmentation, but only some of these result from changes in structural variation or local DNA copy number of the cancer cell genome. Machine learning was used to classify the whole set of fragments as representing either cancerous or normal, and was also used to identify the tissue of origin of the tumor in most cases where there was a cancer. This research shows that a genome-wide diagnostic test could reliably identify most individuals with cancer.
During apoptotic programmed cell death in normal blood cells, as well as cancer cells, nuclease enzymes shred the entire genome. Both DNA strands are cut between the nucleosomes to generate characteristically sized fragments.2 A small proportion of this DNA then floats free in the plasma, presenting only a few copies of each oncogenic mutation. This, against a constant background of DNA from normal blood cells, limits the sensitivity and specificity of diagnostic tests that read cell-free DNA, gene-by-gene.
In contrast, whole genome sequence gives more signals to work with (for example, see Ref. 3 Fig 3B). This is because the position of the nucleosomes of the chromatin of the whole genome influences the fragmentation patterns of cell-free DNA. Nucleosome positions are sensitive to all cancer-associated genomic processes. The differential signal of the test therefore reflects not only mutations and chromosome rearrangements but gene expression changes and even epigenetic alterations such as DNA methylation and histone modification.
1Cristiano S, Leal A, Phallen J, Fiksel J. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019: 570; 385–389
2Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 1980: 284; 555–556.
3Underhill, HR et al. Fragment length of circulating tumor DNA. PLoS Genet 2016 12: e100616.