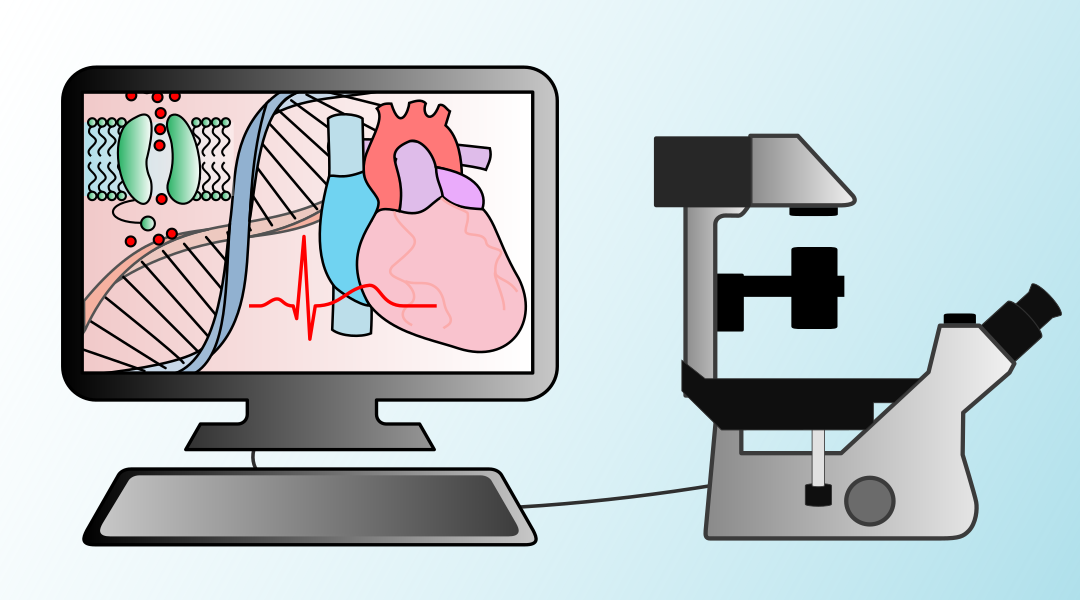
Computer models of the heart’s electrical activity with improved predictive power are being developed through tailored experiments and smarter ways of using data.
Each heartbeat is coordinated by the spread of electrical impulses through the heart. When electrical activity of the heart is compromised, irregular rhythms called arrhythmias can develop, which pose a serious risk of death.
The risk of this happening can be increased by genetic mutations, certain pharmaceutical drugs, the normal aging process, diseases, or combinations of these factors. Preventing sudden death from arrhythmias is a major challenge for healthcare providers the world over. However, understanding how behaviors at the whole-heart level emerge from multiple interacting biological components, also known as the systems biology approach, remains a significant challenge.
Computational and mathematical models are central to the systems biology way of thinking and are now being used by labs across the world to aid cardiac research. Since the first computer model of the heart’s electrical activity was developed sixty years ago, many models have been created ranging in complexity and scale, spanning from the nanoscale “protein” level to the whole-heart “organ” level.
These models are created by studying particular aspects of physiological function in detail and then finding equations that explain (or at least reproduce) experimental observations. Such models can be used to visualize processes that cannot easily be measured during an experiment or are not available to doctors, and to extrapolate to new situations — for example, to predict the response to drugs.
Exciting recent advances include using computer models to reproduce the electrical activity of specific, individual cells; translate findings from one species or cell type to another; and for pharmaceutical safety assessment. Important steps are also being taken towards patient-specific treatments guided by a personalized model of each patient’s heart.
The usefulness of a model in these situations depends on how it was fitted to data (i.e., calibrated), which involves matching model outputs to experimental or clinical observations to estimate parameter values — the unknown numbers in the equations.
This can be done using experimental datasets from the literature, but simulations of experiments can also be used to design better experimental procedures that are guaranteed to provide the information needed for calibration. The quality-of-fit achieved in the final calibration process is sometimes used to assess the model’s performance, but a more stringent method to validate the model is to compare its predictions in new situations, using datasets that were not used in the calibration procedure.
In other words, the way in which these heart models are built is key. How well a model can predict new phenomena depends not only on the model itself, but also the appropriateness of the data used to calibrate the model.
Nevertheless, many ways in which models can be improved as tools for cardiac research do not require more complicated models or new experiments, just better use of the experimental data and reproducible descriptions of the calibration process.
Through further advances and improved standards when it comes to model calibration, it is hoped that mathematical and computational models of the heart continue to be at the forefront of systems biology and cardiac research, and may one day revolutionize the way we diagnose and treat patients.
Written by: Dominic Whittaker
Research article found at: D. Whittaker, et al. WIREs Systems Biology and Medicine, 2020, doi.org/10.1002/wsbm.1482