Nature-inspired investigation has been employed throughout science communities in various fields, from a gecko inspired pick and place gripper to 3D printed bioinspired blood vessels, and much more.
Current methods to design proteins and peptides with desired properties, via computation means, have limitations associated with the type of input information that is used, and therefore the results obtained. A novel, nature template-based computation design method has been proposed, and demonstrates the potential for the design of peptides for chemical biology and medicinal chemistry.

The concept of the hydrophobic moment transferred to 3D space for Magainin-2. See more here.
Pharmacological peptide research is experiencing an apparent resurgence in the discovery of host-defense antimicrobial peptides (AMPs). AMPs were therefore chosen by Gisbert Schneider and co-workers of ETH Zürich and the University of Salzburg as their target for this computational investigation.
They employed their approach for structure-based peptide representation rational in silico design for the generation of AMPs. The design principle generated peptides with desired properties. The first-in-class AMPs were designed and exhibited a defined mechanism of antibacterial action. These results demonstrate that the nature template-based method that they used has practical applicability.
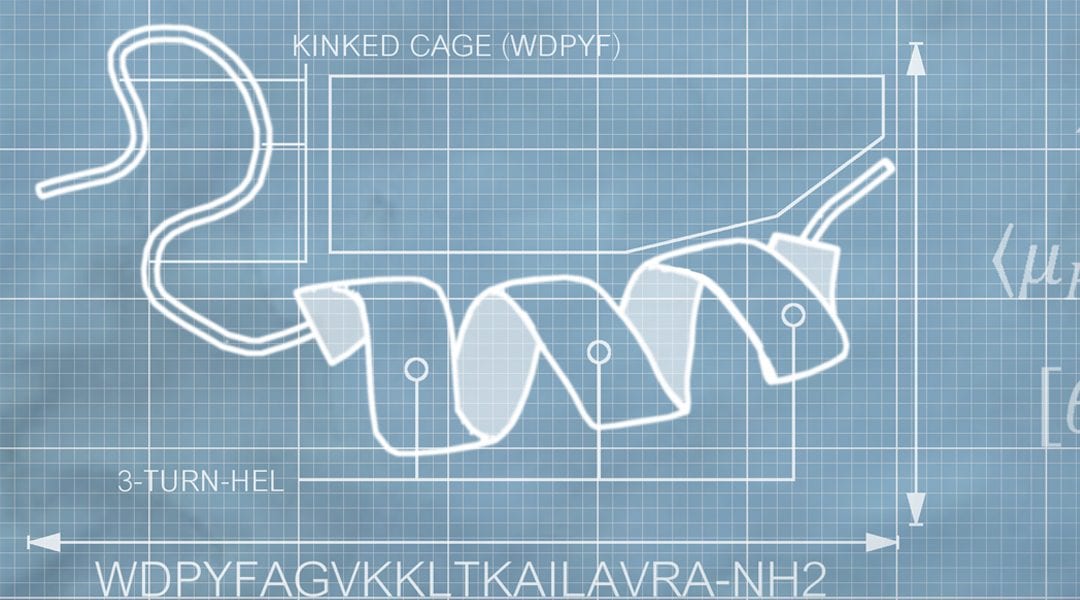
Limitations arising from methods based on residue patterns and physiochemical properties in other computation methods have been overcome with the development of this new method by Prof. Schneider and co-workers. By using 3D structural information and physiochemical properties an interpretable visualization of features was accessible. This enabled key segments to be identified immediately.
The novel peptides that were generated outperformed their natural templates and opens up pathways for further investigation. This novel, nature template-based method is not limited to AMP design and has applicability for the generation of designs with other peptide activities in combination with various amino acid representations.
To find out more, this research article was recently published in Small Issue 40, 2017 as the front cover feature.