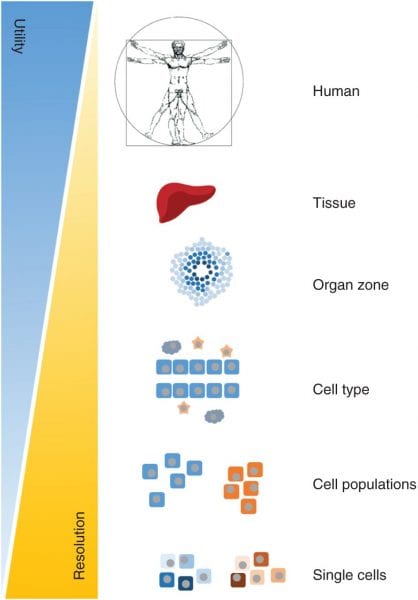
Genome-scale metabolic models (GEMs) are relatively new tools for studying the metabolic changes associated with many diseases. GEMs are reconstructions of the metabolic networks of cells, sometimes able to represent an organism’s whole body. Using GEMs, researchers can analyze metabolic responses and simulations throughout a specific tissue or organ at a particular time in a precise functional state.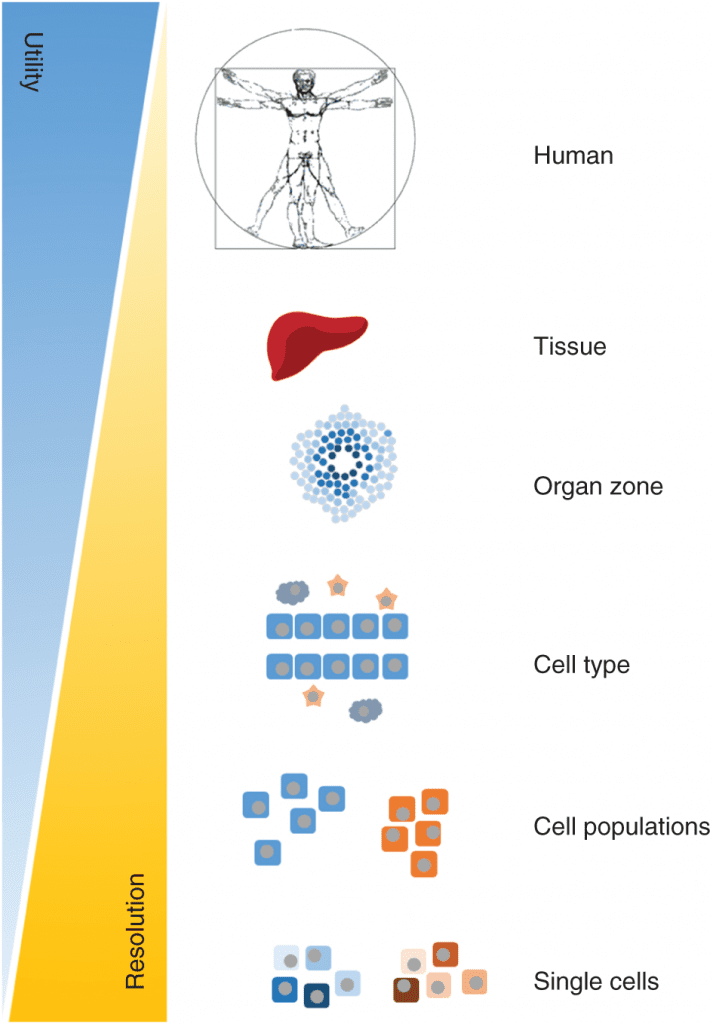
Nutrients enter the metabolic system and are converted to energy, biomass, and byproducts. This process forms a comprehensive biological network. Cell metabolism states are changing properties depending on what nutrients are available, what enzymes are available to act as catalysts, what metabolic fluxes are present, and what enzymes will be available in the near future.
Recent advances in genome sequencing, data accessibility and computational power have contributed to GEMs becoming more popular tools. However, their use for human studies is dependent on the availability of sequenced genomes, the functional mapping of specific genes to metabolic enzymes, and increased computing capacities. Researchers have proven the usefulness of GEMs but still encounter multiple challenges in applying GEMs to human health as well as constructing and simulating them specific organs and diseases.
In this review article “Genome-scale metabolic models applied to human health and disease” from Daniel J. Cook and Jens Nielsen of Chalmers University, Sweden, published in WIREs Systems Biology and Medicine, the authors review these challenges, including building, design and simulation, and look ahead to advances that could further the understanding of the human metabolism. However, GEMs are somewhat limited in terms of the metabolic state’s complexity. Often GEMs do not consider transcriptional or translational feedback.
Researchers are exploring several approaches to address these challenges, such as using proteomic and transcriptomic-informed methods to build GEMs for individual organs, diseases, and patients and using constraints on model behavior during simulation to match observed metabolic fluxes. GEM models are snapshots of the metabolic system at a given transcriptional and proteomic state. Although this review focuses on GEMs application to human health, the challenges and perspectives discussed are also applicable to GEMs use in other fields.
Even given these limitations, GEMs have proven their usefulness for human health in multiple cell types including adipoctyes, macrophages, and microbes in the human gut, and in multiple disease states, including type 2 diabetes, fatty liver disease, and many cancers. Researchers have also built GEMs to describe the metabolism of cancers in individual patients, a step toward patient-specific, targeted treatments.