RNAs are important, long-chain polymeric macromolecules with a range of different cellular tasks. They are chemical copies of the genetic information held in DNA and can serve as templates for protein production (mRNAs), be the structural and/or catalytic components of large molecular macromachines (rRNAs, snRNAs, tRNAs), or regulate gene expression (for example miRNA). The Achilles’ heel of an RNA polymer are its ends—known as the 5´- and 3´-ends. Unprotected ends lead to uncontrolled degradation by enzymes called ribonucleases. To avoid degradation from the 5´-end, for example, most RNAs are protected by a chemical modification. Several different types of such modification exist, but common to all is the presence of at least one pyrophosphate bond.
Importantly, these chemical modifications at the RNA 5´ end need to be reversible to allow RNA turnover, for example to flexibly adapt gene expression to a changing environment. RNA polymers can be degraded sequentially either in the 5´-3´ or 3´-5´ direction. In both cases, the protective pyrophosphate bond needs to be broken, either at the beginning or at the end of the decay process, respectively.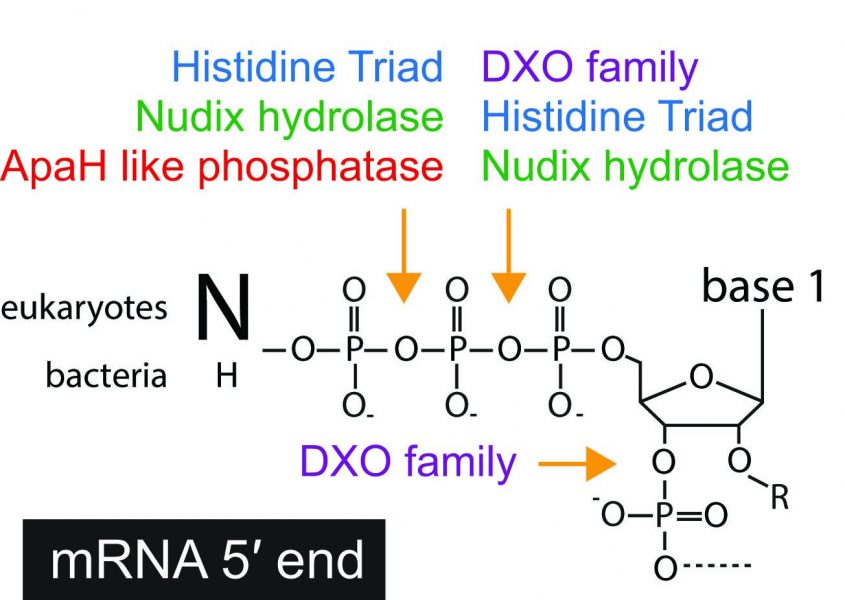
Surprisingly, even though all enzymes involved in the cleavage of RNA 5´ ends share the same activity, a pyrophosphohydrolase activity, they belong to four different enzyme classes. A WIREs RNA Review by Susanne Kramer and Alexander McLennan discusses the differences between these enzymes and how they contribute to RNA decay pathways in eukaryotes and bacteria. In recent years, many novel types of 5´ end modifications have been discovered in RNAs, as well as several novel enzymes involved in the cleavage of these modifications. The most recent and unexpected discovery is a new enzyme from a family known as the ApaH-like phosphatases that removes the rather unusual 5´-end modification from mRNAs in trypanosomatidae, human pathogenic parasites responsible for several neglected tropical diseases. This enzyme could offer a specific therapeutic target in such parasites. It seems we are only beginning to understand the complex mechanisms of RNA 5´ end removal.
Kindly contributed by the Authors.