As a signal for gene-specific silencing of expression in many organisms, double-stranded RNA (dsRNA) is specifically recognized by dsRNA-binding proteins (dsRBPs). Not surprisingly, the dsRBP family controls RNA stability, editing, and function in all eukaryotes. The central feature of this family is the recognition of a generic RNA duplex using highly conserved dsRNA-binding domain (dsRBD) that recognizes the characteristic features of the RNA helix. Variations on this theme that confer species and functional specificities have been reported, but most dsRBPs retain their capacity to bind generic dsRNA.
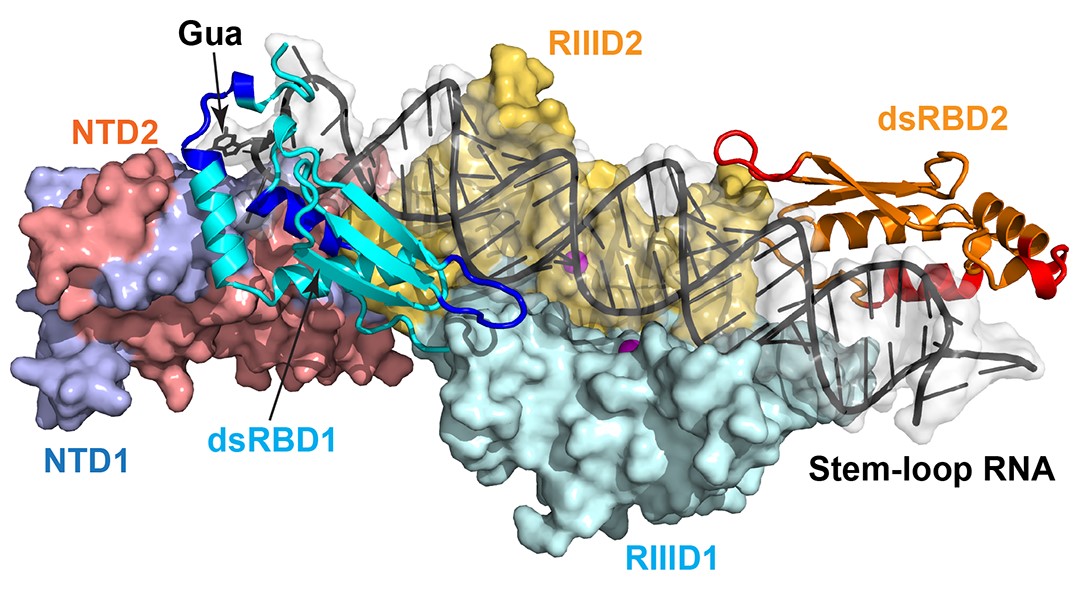
Among dsRBPs, prokaryotic ribonuclease III (RNase III) and eukaryotic Rnt1p, Drosha, and Dicer represent four distinct classes of the RNase III family. Like all dsRBPs and most RNase IIIs, Rnt1p from Saccharomyces cerevisiae has a dsRBD, but unlike most of its kin, it poorly binds to generic RNA helices. Instead, Rnt1p, the only known RNase III expressed in S. cerevisiae that lacks the RNA interference (RNAi) machinery, recognizes a specific class of stem-loop RNAs. To recognize the specific substrates, the dsRBD of Rnt1p is specialized, featuring an unusual abbbaaa topology (in contrast to the canonical abbba dsRBD fold) and a sequence-specific RNA-binding motif at the C terminus of dsRBD. The genetics, function, structure, and mechanism of Rnt1p are described by Sherif Abou Elela and Xinhua Ji in WIREs RNA.
Since the discovery of Rnt1p in 1996, significant progress has been made in studies of its genetics, function, structure, and mechanism of action, explaining the reasons and mechanisms for the increased specificity of this enzyme and its impact on the mechanism of RNA degradation. As the first three-dimensional structure of eukaryotic RNase IIIs in complex with dsRNA in a catalytically meaningful way, the high-resolution crystal structure of yeast Rnt1p post-cleavage complex reveals a double-ruler mechanism for substrate selection and cleavage. The structure shows that Rnt1p recognizes a guanine nucleotide by a G-clamp motif at the C terminus of dsRBD, that the guanine base is also recognized by the N-terminal domain (NTD) of the enzyme, that the G-clamp is required for substrate selection and cleavage, and that the NTD is required for the accuracy of processing. The structure also describes the detailed architecture of the RNA cleavage assembly, explaining how eukaryotic RNase IIIs follow the basic two-Mg2+-ion catalytic mechanism that was first revealed by bacterial RNase III structures. Along with the structures of bacterial RNase III:RNA complexes, the Rnt1p:RNA structure demonstrates that substrate selection by RNase IIIs is accomplished independent of cleavage, allowing all members of the family to follow the same basic two-Mg2+-ion catalytic mechanism for dsRNA processing.
Kindly contributed by the authors.