Proper post-transcriptional regulation is essential for all life on Earth. Over the course of its lifespan, every RNA molecule undergoes dozens of different processing events necessary for proper gene expression. Understanding the regulation of RNA processing reveals fundamental features of gene expression across the eukaryotic domain of life.
In a recent WIREs RNA review, Foley et al. from the University of Pennsylvania discuss the recent explosion of data in the field of post-transcriptional regulation in the model plant Arabidopsis thaliana. Their review discusses the impact of both RNA binding proteins and RNA secondary structure on post-transcriptional regulation. Over the past ten years, techniques have been developed to globally identify RNA binding proteins, and to determine where these proteins bind across the transcriptome. These studies have resulted in a deeper understanding and demonstrated an overall importance of the role of RNA processing in proper flowering timing and circadian rhythm regulation.
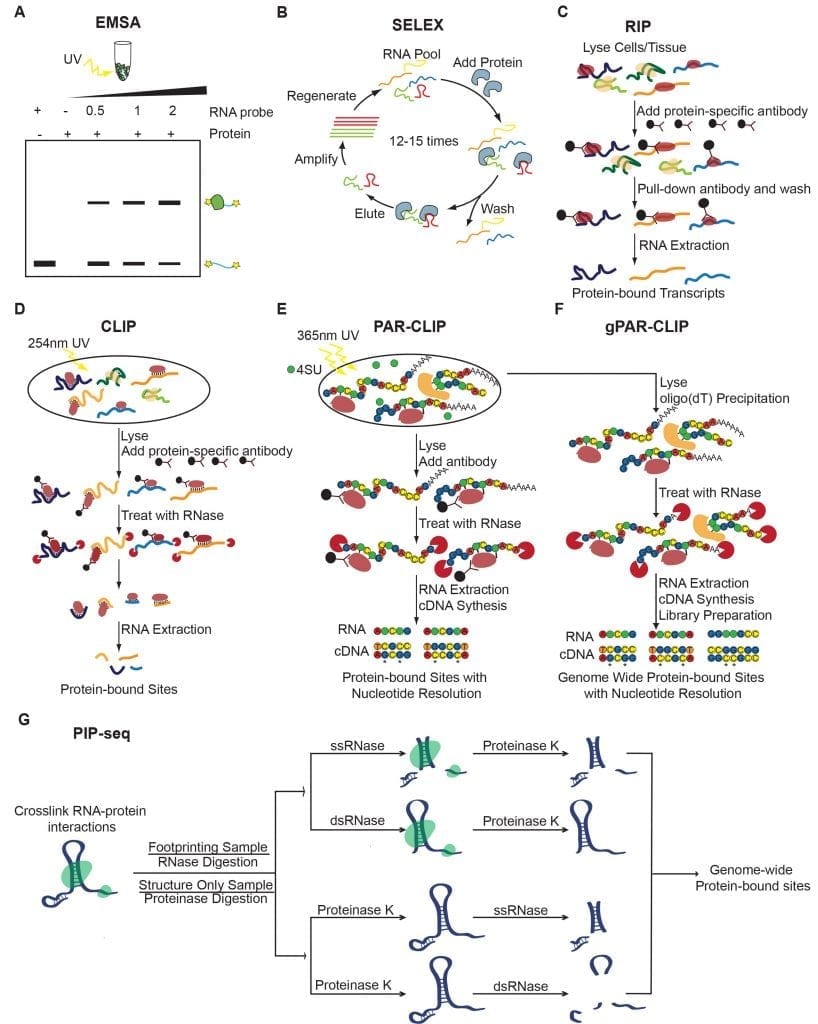
An overview of techniques used to interrogate in vitro, in vivo, and genome-wide techniques used to study RNA-protein interactions. (A-B) In vitro techniques. (A) Electrophoretic mobility shift assays (EMSAs). (B) Systematic evolution of ligands by exponential enrichment (SELEX). (C-E) Target-specific in vivo techniques. (C) RNA immunoprecipitation (RIP). RIP can be followed by RT-qPCR (RIP-qPCR), microarray (RIP-chip), or RNA sequencing (RIP-seq). (D) Crosslinking followed by Immunoprecipitation (CLIP). CLIP can be followed by RT-qPCR (CLIP-qPCR), microarray (CLIP-chip), or RNA sequencing (CLIP-seq). (E) Photoactivatable ribonucleoside enhanced crosslinking and immunoprecipitation (PAR-CLIP). (F-G) Genome-wide techniques. (F) Global photoactivatable ribonucleoside enhanced crosslinking and immunoprecipitation (gPAR-CLIP). (G) Protein interaction profile sequencing (PIP-seq).
Furthermore, recent work has been performed to better understand the role of secondary structure in RNA processing. Each RNA molecule is able to undergo intramolecular basepairing, resulting in single- and double-stranded regions of the transcript. RNA secondary structure has been shown to influence RNA-protein interactions, therefore this feature is an equally important regulator of RNA processing. The field of RNA secondary structure has grown recently, with techniques developed to probe RNA folding both in vitro and in vivo. Foley et al. examines the advantages and drawbacks to each of these techniques, while discussing the major advances made in understanding RNA secondary structure in plant biology. Together, this review provides a survey of the available techniques and fundamental insights into post-transcriptional regulation in plants.
Text contributed by the authors.
Homepage (featured) image credit: Gio.tto/Shutterstock.