In a normal cellular context, the majority of the DNA is present in a defined conformation—the so-called B-form. This conformation represents a right-handed double helix with 10 to 10.5 bases per turn. In addition to this B-form, the DNA can also adopt the so-called A-form—a right-handed double helix with a more compact structure than the B-form; and the Z-form—a left-handed helix. Furthermore, when opposite DNA strands are paired in a non-sequential manner, other non-B-DNA structures such as single-stranded DNA, hairpins, and triplex and quadruplex structures may form.
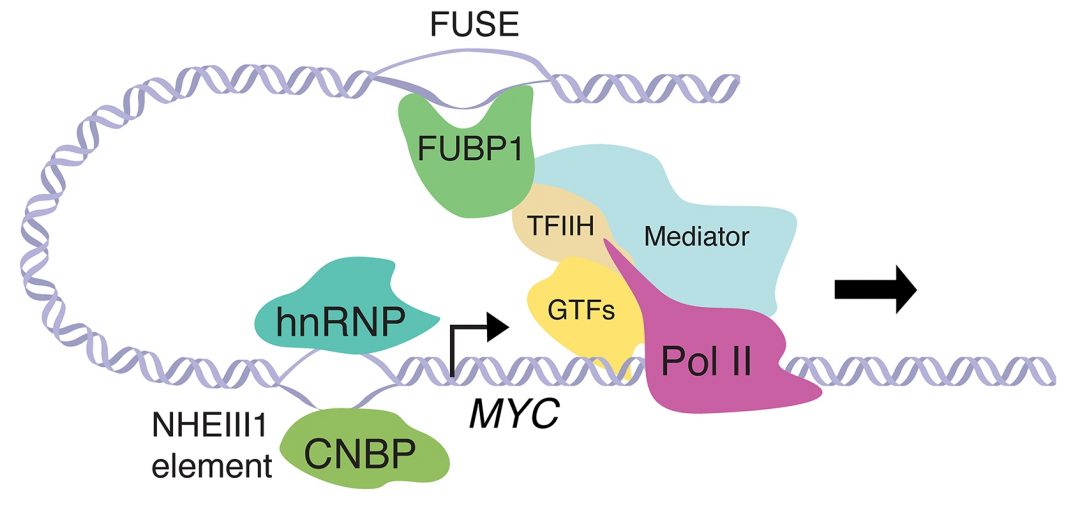
Taking the MYC promoter as an example, Olga Zaytseva and Leonie Quinn discuss the influence of these different DNA conformations on gene expression. MYC is a transcription factor of the basic helix-loop-helix family and a potent oncogene: Elevated levels of the protein promote tumorigenesis by activating genes required for cell growth and proliferation. Therefore, transcription of MYC needs to be tightly regulated.
In general, transcription generates DNA supercoiling, and this in turn impacts DNA topology and provides feedback mechanisms. The twin-supercoiled-domain model developed in the late 1980s posits that positive supercoiling waves are generated ahead of the RNA polymerase, whereas negative supercoiling is found behind the transcription machinery (see Figure below).
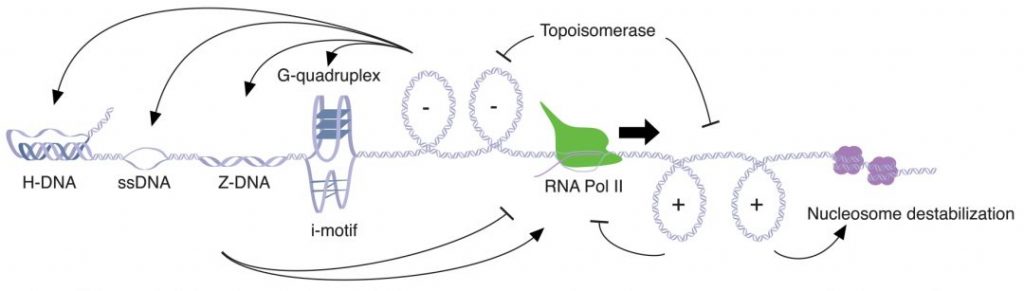
In the case of MYC, a specific region called NHE III1 (nuclease hypersensitive element III1) exists in a dynamic equilibrium between double-stranded DNA and secondary structures induced by supercoiling. These secondary structures include the G-quadruplex, i-motif, H-DNA, and single-stranded DNA, and together provide regulatory feedback for transcriptional regulation of MYC.
Recent genome-wide studies indicate that the supercoiling mechanisms described above are not restricted to the MYC promoter, but influence gene expression of many or maybe even all genes.
Positive supercoiling found ahead of the RNA polymerase reduces the spacing between turns in the double helix, thus increasing the rigidity of the DNA. When this supercoiling is not resolved by the action of topoisomerases, it may lead to a block in transcription. Negative supercoiling on the other hand leads to DNA that is more likely to melt into single-stranded DNA and to form non-B-DNA structures. It is therefore not surprising that highly transcribed genes, such as the ribosomal RNA genes, display negative supercoiling.
With these genome-wide findings, DNA structural conformation has emerged as another important player in the regulation of gene expression.