RNA is a hugely versatile molecule with roles ranging from enzymatic, to regulatory to structural functions, to serving as the blueprint for protein synthesis, and as storage molecule of genetic information in viruses.
It is a well-known fact that RNA molecules are subject to posttranscriptional modification including the attachment of diverse chemical groups to its bases and the ribose moiety. The majority of these modifications are methylations.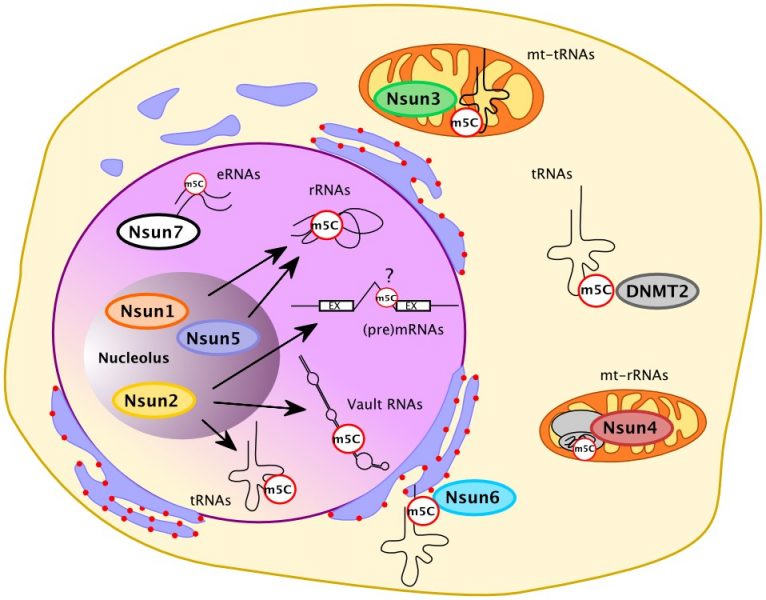
Although RNA modifications have been discovered half a century ago, research on it has focused on the abundant classes of RNA, the rRNA, and tRNAs. The major advancements of the past few years in the development of methodologies for the detection of RNA modifications has reignited the interest in RNA modifications in particular also in the context of less abundant types, such as mRNA or long non-coding RNAs and has led to the coining of the term “epitranscriptomics” to describe this field of research.
As detailed in a WIRES RNA review article by Trixl and Lusser of the Medical University of Innsbruck, Austria, methylation of cytosine, which is a modification typically occurring in DNA where it is considered an “epigenetic” mark, has recently also been characterized in various types of RNA.
Although the currently used methods for 5-methylcytosine (m5C) mapping in the transcriptome are not free of technical problems, the distribution of m5C in mRNAs revealed enrichment at specific locations, such as the 5’ and 3’ UTRs.
While the biological significance of m5C in mRNA is still rather obscure and may involve effects on nuclear cytoplasmic shuttling of RNA or on translation, cytosine methylation of tRNAs can serve to change tRNA structure, integrity, and decoding potential and seems to be particularly important under stress conditions. There is a number of RNA cytosine methyltransferases whose known targets are predominantly tRNA and rRNA. Only one, Nsun2, which is known to cause developmental defects when mutated and is overexpressed in a number of cancers, has to date been implicated in methylating other RNA types. Although knowledge about m5C in RNA is just beginning to consolidate, cytosine methylation emerges as a powerful mechanistic strategy to regulate cellular processes on an epitranscriptomic level.
Kindly Contributed by the Authors.