Ribosomes are molecular machines dedicated to the synthesis of proteins. They comprise two subunits, a large and a small, respectively called in eukaryotes the 40S and the 60S subunit. Both subunits feature an RNA backbone within which numerous ribosomal proteins (RPs) are integrated. Producing ribosomes is one of the major tasks of any cell. This production is regulated depending on the metabolic state of the cell. It requires the intervention of the three RNA polymerase machineries and hundreds of accessory assembly or maturation factors (AFs).
Synthesis of ribosomes starts in a specific nuclear compartment termed the nucleolus, in which transcription of precursors to the mature ribosomal RNAs (rRNAs) takes place. During the transcription process, these precursors start assembling with a subset of RPs and AFs to generate a first pre-ribosomal particle that quickly gives rise to the first pre-40S and pre-60S pre-ribosomal particles, precursors to the mature small and large ribosomal subunits, respectively. These particles then follow independent maturation pathways in the nucleolus, then in the nucleoplasm and finally in the cytoplasm, after an export step through the nuclear pores. By “maturation,” we mean the trimming of pre-ribosomal RNAs (pre-rRNAs) to their mature size by ribonuclease enzymes, their chemical modification, their correct association with RPs, and their adoption of a correct three dimensional fold. The latter aspect is crucial since the activity of the ribosome is mostly carried by its RNA constituents, that properly function only if correctly folded.
In WIREs RNA, Emilie Cerezo and colleagues review the current knowledge of the maturation process of pre-40S particles in both yeast and human cells. It highlights the information gained from the recently published three dimensional atomic structures of cytoplasmic yeast and human pre-40S particles. Overall these human and yeast structures are similar and point to two fundamental roles of the core AFs. They occlude the binding sites on rRNAs of translation factors and in doing so, they prevent untimely entry of 40S subunit precursors in the translation (i.e., protein producing) process, thus avoiding potentially extremely deleterious translation errors. They also maintain, while they are bound to pre-40S particles, certain rRNA helices in specific configurations that presumably promote both the correct association of RPs and the stepwise adoption by the rRNAs of their correct three dimensional fold.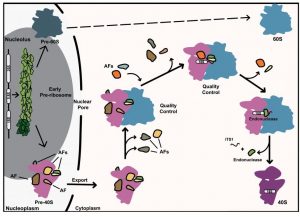
This WIREs RNA review nevertheless stresses two major differences between the yeast and the human pre-40S maturation processes. The processing steps that trim the human 18S rRNA precursors to generate the mature 18S rRNA, the rRNA component of the 40S ribosomal subunit, involve a much more complex array of diverse enzymes (including ribonucleases and poly (A) polymerases) than in yeast. The biological rationale for the mobilization of such a complex enzymatic machinery may be linked to the longer size of the sequence to be removed in the 18S rRNA precursors in humans compared to yeast, but still needs to be further explored.
The second major difference is the discovery in yeast of a proofreading mechanism of cytoplasmic pre-40S particles. This proofreading system probes the ability of the pre-40S particle to associate with a 60S ribosomal subunit and a subset of translation factors, before the last maturation steps that release the mature 40S subunit competent for translation are carried out. Aberrant pre-40S particles that fail this proofreading step are presumably destroyed by as yet unknown degradation pathways. No such proofreading system has been described so far in humans but it is highly probable that other modes of quality control exist in humans, potentially even more sophisticated. Discovering these human quality controls constitutes an exciting challenge for future research in the field.
Kindly contributed by the Authors.
Adapted featured image license information: Original ribosome by David S. Goodsell and the RCSB PDB for Molecule of the Month–January 2010 under the CC-BY-4.0 license.