There are thousands of different RNA molecules in every cell that perform a wide range of roles. Some RNAs carry information to make proteins, others regulate cellular processes, while still others are degraded within minutes of being made. This incredibly dynamic population of RNAs is regulated both by how quickly they are made (synthesis) as well as how quickly they are degraded, and these rates can vary widely for different RNA transcripts.
This juggling act between synthesis and degradation allows cells to rapidly alter RNA levels by tuning one or both of these parameters in response to the cell’s environment. Despite evidence that RNA dynamics control many aspects of cell metabolism, methods to measure RNA levels via high-throughput sequencing (RNA-seq) tend to provide a static perspective on cellular RNA populations, because there is no way to distinguish newly made RNAs from old ones.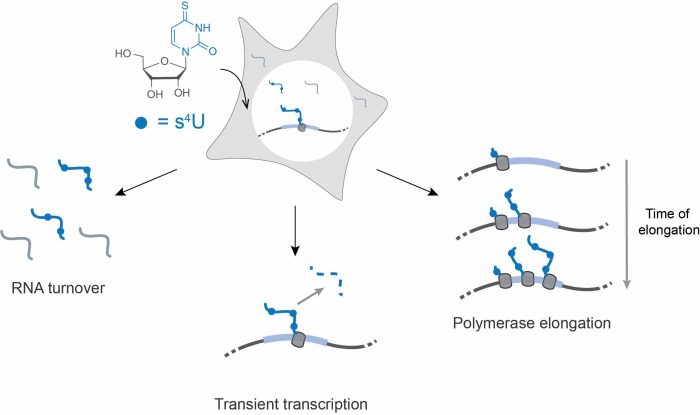
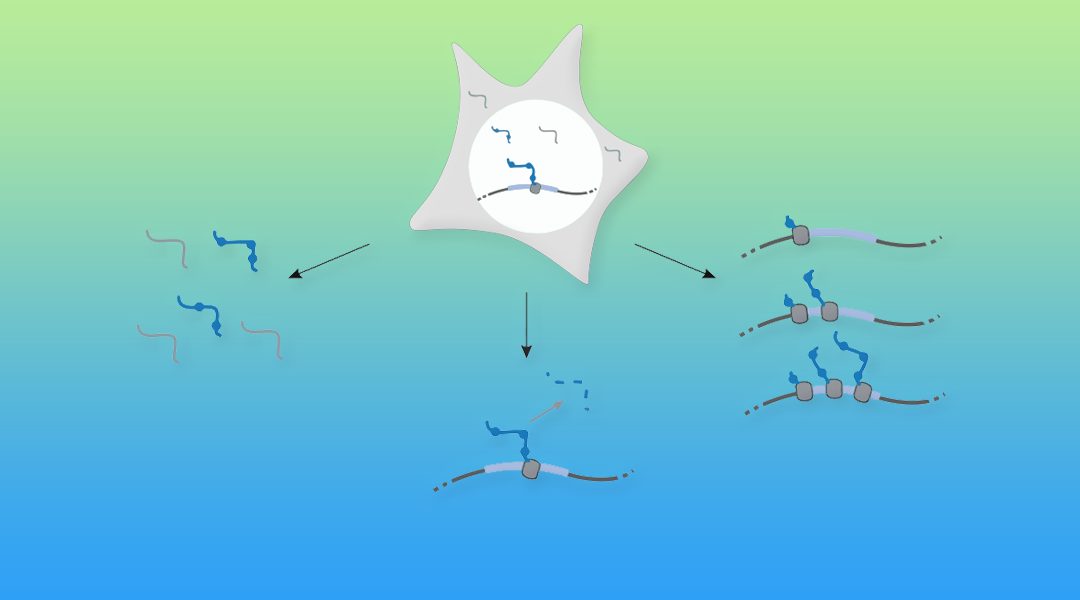
To study these dynamics, cells can be fed RNA building blocks (nucleosides) that contain chemical groups that are rare or absent in cellular RNA, and these nucleosides are incorporated into newly made RNAs. The chemical groups are chosen to be largely invisible to the cell, yet the chemical properties of the non-native nucleoside can be leveraged in order to separate the two populations of RNA: the pre-existing, unlabeled fraction, and the newly made, labeled fraction.
In WIREs RNA, Erin Duffy, Jeremy Schofield, and Matthew Simon review methodologies and also important considerations of using a specific metabolic label, 4-thiouridine, to probe RNA population dynamics.
This chemical strategy, combined with high-throughput sequencing technology, has promoted an explosion of diverse methodologies to study different aspects of RNA population dynamics. For instance, rates of RNA synthesis and degradation can be measured for cells at steady-state.
Alternatively, rare RNAs that are rapidly degraded and therefore difficult to detect in standard RNA-seq, termed transient transcripts, can be enriched from the total RNA pool for more sensitive detection. In addition, when cells respond to an environmental stimulus, metabolic labeling can distinguish between mechanisms that regulate the cellular response (i.e., changing the rate of RNA synthesis versus degradation). These experiments have been applied to yeast, cell culture, and even mammalian tissues.
Recent improvements in chemistry, sequencing technology, and bioinformatic analysis offer a bright future for understanding the biology of RNA population dynamics.
Kindly contributed by the Authors.