Viruses are highly ordered supramolecular complexes that infect cells in all kingdoms of life. From a physicochemical perspective, they can be regarded as molecular machines that have successfully evolved to spread between related organisms by hijacking the host cell’s machinery. Although viruses are very diverse, they share common functions and properties, such as the ability to form hollow protein shells, named capsids, to protect their genome from the external environment.
Viral capsids have functions that go beyond genome protection, as they play key roles in signalling and regulation during the viral life cycle. Therefore, capsid formation and capsid-genome interaction, together with capsid stability, are crucial features of viruses and are instrumental to understanding the viral infection. In addition, fundamental viral mechanisms are of growing importance in disciplines such as biomedicine and (bio)nanotechnology, e.g., using viral particles as vehicle for targeted drug delivery. However, the detailed mechanisms involved in capsid formation and their related meta-stabilities remain poorly understood. But by using physics-based techniques, this can be unravelled.
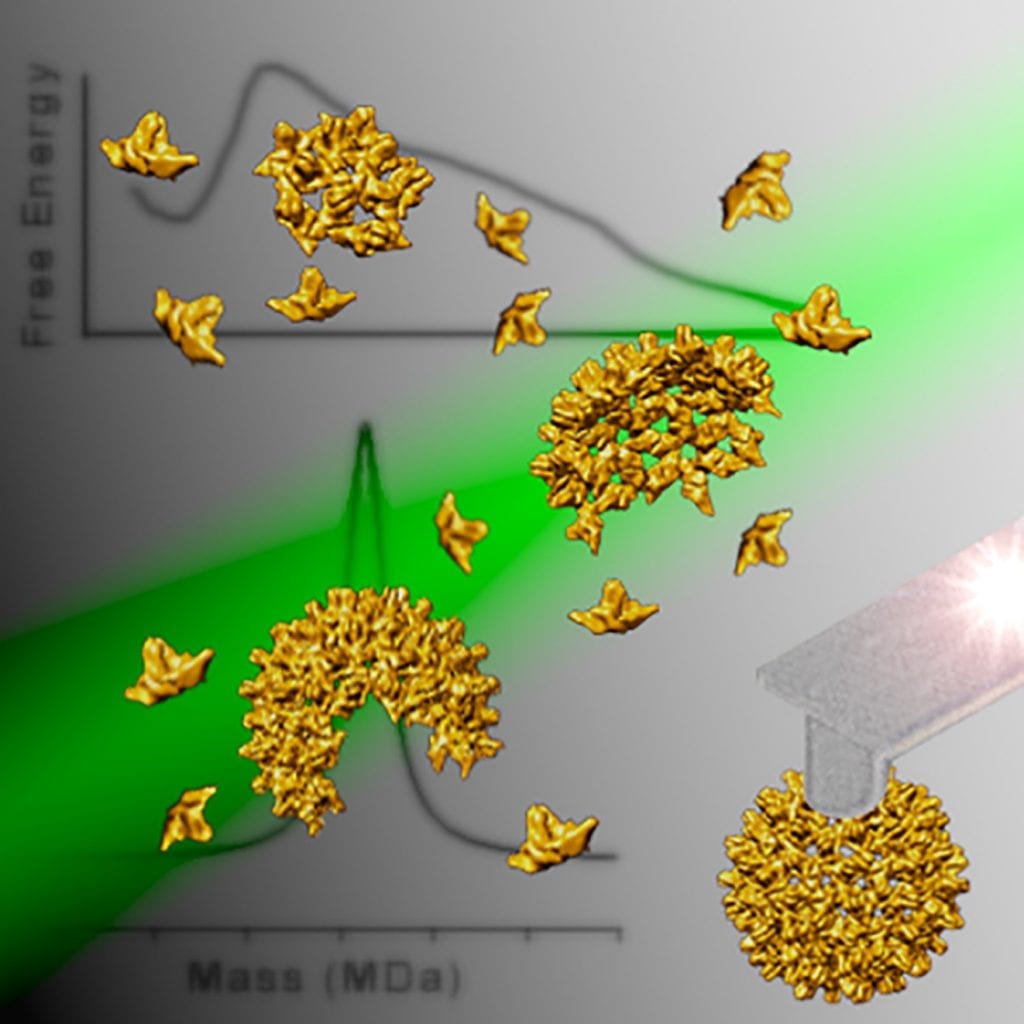
In a recent study published in WIREs Nanomedicine & Nanobiotechnology, Dr. Wouter Roos and co-workers from the Rijksuniversiteit Groningen in the Netherlands explore physics-based studies of viral systems and the recently fast-growing field of physical virology.
Capsid formation can occur through the self-assembly of all the viral components or by actively packaging genome using a molecular motor after a pro-capsid has been formed.
“In the simplest scenario of capsid formation, the functional capacity to self-assemble resides in the primary amino-acid sequence of the capsid proteins and, hence, the folded structure of the viral protein subunits. Thus, the assembly process is solely driven by protein-protein and, for co-assembly with viral nucleic acids, protein-genome interactions,” according to the authors in their study. “The balancing of these interactions…, implicitly conditioned by the surrounding environment, will ultimately determine the assembly pathway(s).”
Indeed, different self-assembly pathways are allowed: some viruses form a specific nucleation structure from which they grow to full capsids, while other viruses form amorphous protein complexes around the genome that then relaxed to a symmetrical capsid; viruses can encode specific sequences in their genomes that assist the assembly or packaging, named packaging signal; other viruses instead do not show clear genome specificity signs where viral genome topology seems to play a key role.
“Overall, it seems that there is a correlation between the Young’s modulus and the manner of encapsidation of viruses. While viruses that self-assemble around their genome possess a relatively low Young’s modulus, the modulus is higher for particles that self-assemble empty and that use a packaging motor to encapsidate the genome,” said the authors.
“In addition, viral systems have been proven to be highly dynamic, showing the capability to modulate stability during infection, which highlights their remarkable conformational plasticity and adaptability.” The viral life cycle is indeed composed of many stages, and viral capsids have to adapt their mechanical properties to the environment of every step to pursue their main goal to facilitate virus propagation.
“The hereby newly generated insights will likely not only advance fundamental science, but also applications of viruses and virus-like particles.”
Kindly contributed by the authors