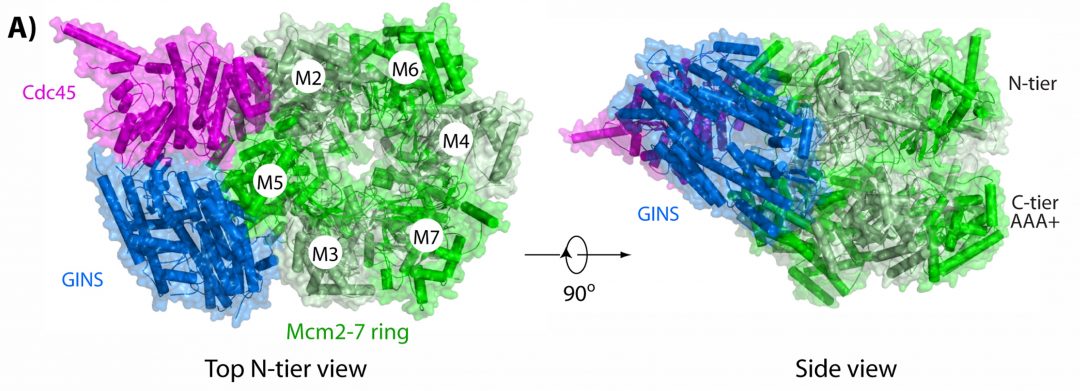
The eukaryotic CMG helicase is a multi-subunit complex containing Cdc45, an Mcm2-7 ring that encircles DNA and the GINS tetramer. First, during G1 phase of the cell cycle, two Mcm2-7 rings are loaded onto an origin of replication. Subsequently, in S phase, Cdc45 and GINS are assembled onto each Mcm2-7 ring. This then results in two active CMG helicases enabling the formation of replications forks that travel in opposite directions. The assembled CMG helicases also act as a central scaffold for overall replisome organization recruiting other proteins necessary for DNA replication such as DNA polymerases and other factors.
In their review published in BioEssays, Huilin Li and Michael O’Donnell take a detailed look at how the subunits are assembled onto DNA and how the resulting complex then moves along the DNA. Interestingly, when assembled onto an origin of replication in the way described above, the two CMGs are directed inward, toward one another in a head-to-head configuration. This, however, implies that each CMG must pass the other in order to leave the origin, and the authors propose a model on how this might be executed.
Once replication is initiated, synthesis of the leading strand requires Pol ε while replication of the lagging strand is performed by Pol α-primase. This is achieved by Pol ε and Pol α-primase binding on opposite faces of CMG forming a highly asymmetric structure. Taking into account recent high resolution electron microscopy studies, Li and O’Donnell review impressive details about how CMG engages the DNA fork and how this complex moves along the DNA. CMG is able to adopt two different conformers: a compact and an extended conformation. Conformational changes are supposedly driven by ATP binding and hydrolysis, and this seems to lead to an inchworm driven motion along the DNA.
While these recent structural studies using different electron microscopy techniques have already led to a better understanding of the events at the replication fork, the authors also point out some open questions and call for the application of new techniques in addition to these structural studies to solve these open questions.