 Traditionally, polymer chemistry has always been the science of “polydispersity” and “distribution”. This situation has gradually changed during the last fifty years. There is indeed a continuous search for better methods to master macromolecular structure.
Traditionally, polymer chemistry has always been the science of “polydispersity” and “distribution”. This situation has gradually changed during the last fifty years. There is indeed a continuous search for better methods to master macromolecular structure.
In particular, the discovery of living ionic polymerizations and controlled radical polymerizations has opened up a wide range of opportunities for polymer synthesis. These methods allow preparation of synthetic macromolecules with very low polydispersity index (e.g. 1.01 for anionic polymerization and 1.05 for controlled radical polymerization) and controlled architectures.
The next step in the field is probably the absolute control of macromolecular structure, i.e. the synthesis of perfectly-monodisperse macromolecules. Many groups have aimed to prepare synthetic polymers that compare in quality with natural polymers such as proteins and nucleic acids. Indeed, biopolymers exhibit precisely controlled primary, secondary, tertiary and quaternary structures and are therefore able to perform advanced tasks.
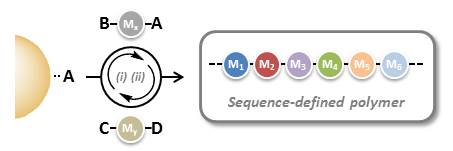
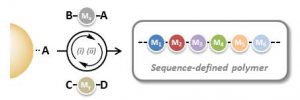
The objective of the emerging field of precision polymer chemistry is to master these structural parameters in non-natural polymers. In particular, the control of comonomer sequences (i.e. primary structure) in synthetic polymers has received increasing academic attention in recent years.
In order to synthesize monodisperse sequence-defined polymers, new synthetic methods have to be identified. Solid-phase iterative chemistry, which has been initially introduced for the synthesis of oligopeptides and oligonucleotides, is an interesting methodology for preparing monodisperse sequence-defined polymers.
However, such approaches are usually time-consuming and request demanding coupling/capping/deprotection steps. Yet, interesting protecting-group-free methodologies have been described in recent years for simplifying and accelerating these processes. In their recent Trend article, Lutz and coworkers describe in details the pros and cons of these new strategies.