In a WIREs Systems Biology and Medicine article, Dejun Lin, Giancarlo Bonora, Galip Gürkan Yardmici, and William S. Noble give an overview of the recently discovered hierarchical three-dimensional (3D) organization of the genome, focusing on computational approaches for elucidating and interpreting this structure. The authors cover a wide range of computational methods that aim to detect and validate 3D organization of the genome from chromosome conformation experiments, reviewing state-of-the-art computational methods in the field, and touching on the relevance of 3D organization with respect to a wide variety of critical biological processes.
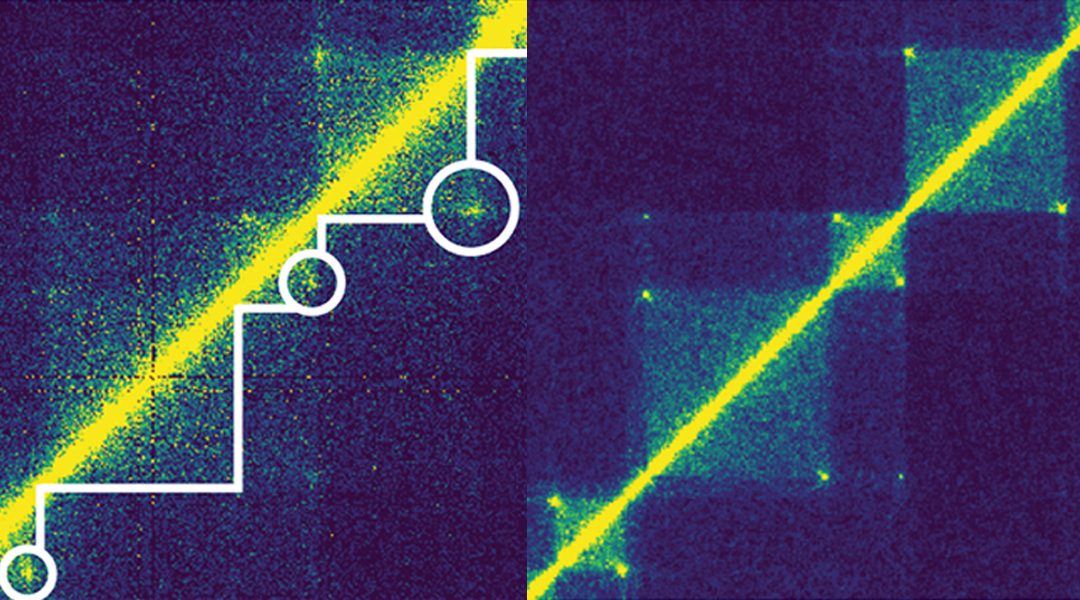
In particular, this WIREs Systems Biology and Medicine article describes the loop extrusion model as an example of how computational approaches can lead to hypothesis generation and validation of putative novel mechanisms in the field of genomics, as shown in the figure. 
Loops are formed by pairs of genomic loci that are spatially close to each other, appearing as peaks in chromosome conformation data that can be detected through computational methods. By investigating the features of DNA around these loops, researchers formulated hypotheses about the putative mechanisms and protein complexes that form and maintain such loops. Through additional experiments and computational simulations, the well-known protein complexes CTCF and cohesin were established as drivers of loop extrusion, which explain many aspects of 3D organization of DNA at a high resolution.
Lin and his colleagues conclude their article by discussing the shortcomings and limitations of current computational approaches, and they speculate on future directions for this important field. For instance, the authors predict that incorporation of imaging data, including super high resolution microscopy and single cell chromosome conformation data will possibly reveal finer features of 3D organization of genomes and deepen our understanding of the variation of genome architecture during many biological processes.
Kindly contributed by the Authors.