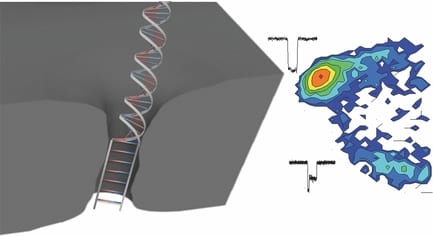
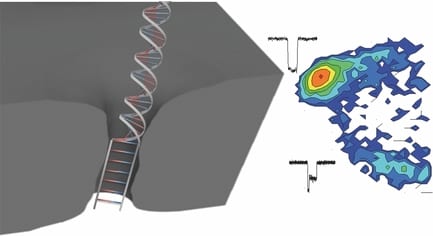
 Nanopore technology is among the most active and promising techniques for third-generation cost-effective, fast, and label-free DNA sequencing. When negatively charged DNA molecules are electrically driven through a tiny pore in a thin membrane in aqueous solution, A, C, T, G signals might be distinguished in real time in ionic current due to different blocking levels to the pore conductance. To achieve this, enough small nanopores with size comparable to the diameter of single stranded DNA (ssDNA) are highly desired in order to ensure single-file passage of DNA and achieve a high detection sensitivity (high signal to noise ratio). Making these sub 2-nm nanopores is feasible by a manually controlled method, such as transmission electron microscope (TEM) based-drilling or ion-beam sculpting, however this is really challenging due to the low yield, complex fabrication procedures, and a labor and time consuming process.
Nanopore technology is among the most active and promising techniques for third-generation cost-effective, fast, and label-free DNA sequencing. When negatively charged DNA molecules are electrically driven through a tiny pore in a thin membrane in aqueous solution, A, C, T, G signals might be distinguished in real time in ionic current due to different blocking levels to the pore conductance. To achieve this, enough small nanopores with size comparable to the diameter of single stranded DNA (ssDNA) are highly desired in order to ensure single-file passage of DNA and achieve a high detection sensitivity (high signal to noise ratio). Making these sub 2-nm nanopores is feasible by a manually controlled method, such as transmission electron microscope (TEM) based-drilling or ion-beam sculpting, however this is really challenging due to the low yield, complex fabrication procedures, and a labor and time consuming process.
Now, researchers from University of Ottawa, Canada have had a smart idea to automatically fabricate 2-nm solid-state nanopores with high yield (>95%) using controlled dielectric breakdown. They applied an electric field across a thin insulating membrane (10 thick SiN) in solution near its dielectric breakdown strength to create a single pore when seeing an abrupt increase in leakage current. A number of nanopores (N=23) have been achieved with an average diameter of 1.3+- 0.6 nm. Several key conditions were optimized in this work in order to achieve a reliable and reproducible automated fabrication, such as putting the fabricated pore into high LiCl solution (pH=8), rapidly turning off the voltage upon finding the first breakdown event, and using enough thin SiN membranes.
These tiny nanopores can serve as super-sensitive DNA single-molecule detectors for not only dsDNA, but also ssDNA. ssDNA and dsDNA are employed as molecule rulers to confirm the nanopore dimension. A longer dwell time was observed for ssDNA in a 2.0 nm nanopore due to an interaction dominated translocation, with a much stronger interaction between ssDNA and nanopores than dsDNA with pores. dsDNA molecules cannot pass through the 2.0 nm nanopore due to their bigger size, instead just showing intercalation at the entrance of the pore, or collisions with the nanopore. In another bigger pore (d=2.4 nm), in addition to observe single file translocation of dsDNA, the authors detect the B-S stretching transition in dsDNA, which is not possible to observe in larger size nanopores (d>5 nm). This transition would uncoil the double helix, resulting in a 1.7 times increase in DNA length. The length increase would decrease its cross-sectional area, therefore leading to a trapped and smaller current blockage in these overstretched two-level events.
In the future, these low-cost, automated fabricated sub-2-nm nanopores with sub-nm precision can be extended to other materials, and can certainly contribute to various applications for personalized genomics and possible DNA sequencing.