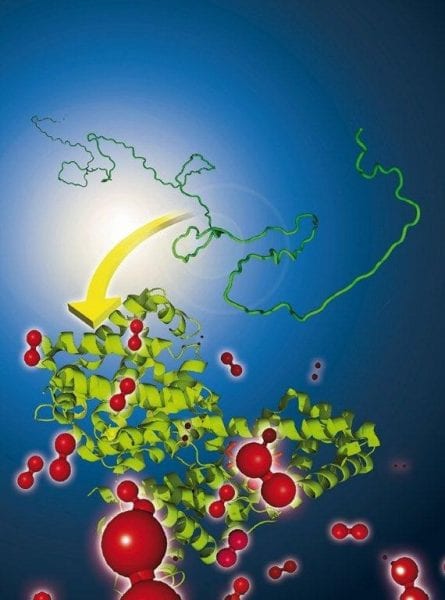
Living subjects are very complex systems and, at the same time, stunningly robust and accommodative. The secret of their success is the proteins which build up the cells of the organisms, which act as cleaners, messengers, transporter, motors, and and much more. How proteins are genetically coded, and how their linear chains of amino acids are put together, is in principle well known. Yet this is not the full story. Before proteins can do their jobs, they have to be folded correctly. This is important because their shape determines the function of the protein, and misfolded proteins can cause severe health disorders such as Alzheimer’s disease – in this case because the proteins seem to aggregate into so-called amyloid plaques.
It is still impossible to predict from the amino acid sequence how the corresponding protein will fold. New insights into the principles of protein folding could enhance the understanding of amyloid-based diseases. In addition, protein drugs could be designed to target specific molecules.
Nobel laureate Ahmed H. Zewail and his co-worker Milo M. Lin summarize their insights into the physical backgrounds of protein folding in the latest issue of Annalen der Physik as part of the renowned Einstein Lectures. This series, established in 2005 in the course of the Einstein Year, comprises articles of reputable scientists such as Nobel laureates Theodor Hänsch, Roy Glauber, and Peter Grünberg. Now, Zewail and Lin report on physical views of protein folding.
Many proteins are able to fold themselves quickly and properly into a complicated conformation without any help of a cellular “folding apparatus”. How they achieve this is still a mystery. Zewail and Lin at the California Institute of Technology in Pasadena (USA) want to solve this “protein folding problem“ by looking at the basic physics behind the complex world of proteins.
Aside from thermodynamics, which control the stability of the possible protein conformers, kinetic aspects play a key role as they rule the mechanisms and time scales of the folding process. A polypeptide chain could spend more time than the age of the universe “trying” all of its possible conformers to find its native one. In reality, folding never takes more than a few seconds.
Using ultrashort laser pulses, Zewail’s team was able to determine how fast the first twist of an alpha-helix is formed. Real-time investigation of chemical reactions with ultrashort laser pulses is one of Zewail’s specialities; his research in this area had been acknowledged with the donation of the Nobel prize in 1999.
The folding of the first helix twist is the fastest step of protein folding; it therefore determines the general speed limit of folding velocity. Without having to know the details of all mechanisms involved, the scientists were able to calculate this general limit with the help of simple analytical models.
The models used are based on a consideration of the torsion angles in the protein backbone plotted in a so called Ramachandran diagram. Sterically forbidden, restricted and free regions are distinguished. In addition, the polypeptide chain was calculated as a three-dimensional lattice thereby arranging the chain in a way that maximizes contacts between hydrophobic residues reflecting their shielding from the physiological aqueous environment. The polar sidegroups, instead, have to point outwards. This corresponds to the natural conformation of proteins.
Based on their calculations, Zewail and his colleague argue that hydrophobic interactions between non-polar amino acid residues are sufficient to fold a polypeptide chain into its native shape in a reasonable time. But this works only if the chain is not longer than ca. 200 amino acids. This could be an explanation for the observation that proteins are built up of domains: Although proteins, in general, may consist of more than 1000 amino acids, they are often composed of domains of independently folding sub-parts. These domains are on average 100 amino acids long, most of them being less than 200. This would correspond to the theoretically determined length regime for which hydrophobic interactions are sufficient for fast folding.