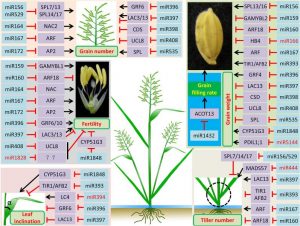
A summary of the miRNAs (highlighted in light blue) and their target genes (highlighted in purple) controlling rice yield‐related agronomic traits, including grain number, grain weight, tiller number, fertility, and leaf inclination. The miRNAs marked in red color are those controlling just one of the above mentioned five traits, based on the recent publications.
Rice, Oryza sativa, is one of the most important food crops in the world. In rice, a number of quantitative trait loci (QTLs), defined as specific locations in the genome, and controlling yield-related traits, have been identified. These loci contain genes which control traits that can be quantified, such as organ height, weight, number and so on.
Some of the genes present are actually microRNA (miRNA) genes, which after expression and processing form 20–24 nucleotide long mature miRNAs—small RNAs which do not form proteins but suppress the expression of target genes involved in regulating various functions related to growth, development, and stress responses in plants and animals.
On the one hand, many miRNAs are found to regulate more than one yield-related trait such as tillering (production of side shoots), grain size, and branch number of a panicle (branched inflorescence bearing flower/grain cluster). However, a rice yield-related trait is usually controlled by multiple miRNAs; for example, grain size is controlled by miR156, miR167, miR396, miR397 and miR1432. In rare cases, a single miRNA may specifically regulate only one yield-related trait, such as miR444, which regulates rice tillering.
In this WIREs RNA review, the authors discuss the role of different miRNAs controlling yield-related quantitative traits in rice, including panicle grain number, grain weight/size, panicle length and branching, tiller number per plant, seed setting rate, and leaf inclination. This knowledge is useful to identify such miRNAs, whose expression can be modulated using modern molecular biology tools.
These approaches involve expressing decoys/mimics of specific miRNAs in plants in the form of target mimics, short tandem target mimics, and sponges. These decoys sequester and/or degrade mature target miRNAs, preventing their accumulation in sufficient amounts to suppress their respective target genes.
To suppress the expression of MIRNA genes or target genes, controlling quantitative traits, RNA knockdown techniques or gene editing tools, such as CRISPR/Cas9, can be used. All constructs targeting miRNAs or their target genes can be expressed using constitutive, ubiquitous, tissue-specific or inducible promoters to achieve silencing in a desired manner, spatially or temporally.
Altogether, modulating the expression of a miRNA or its target gene(s), can alter quantitative traits in rice for crop improvement.

















