The development of a single cell into a living organism depends upon an array of proteins being present at the correct time and in the correct location. These proteins are instrumental in controlling the growth, division, and movement of cells to form a 3-dimensional embryo. The first step in the control of gene expression transcription from RNA from DNA which has been well studied.
However, it is now clear that each step of the post-transcriptional pathway is also vitally important in determining the final levels of gene product. Up until relatively recently, it has generally been assumed that RNA stability plays a passive, rather than active role in the post-transcriptional pathway. However, recent work has shown that it is highly regulated, varies during development or in response to environmental factors, and that specific RNAs can be targeted for degradation.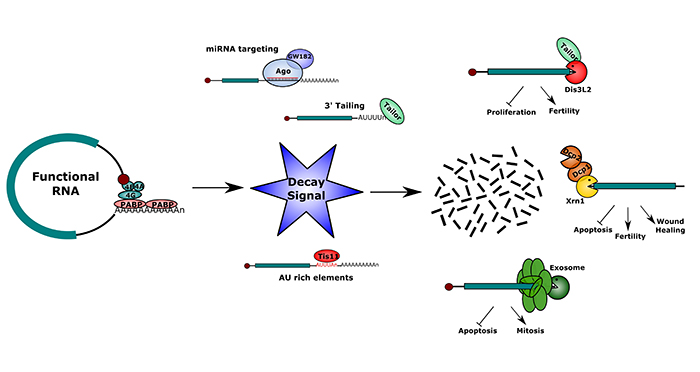
In WIREs RNA, authors Benjamin Towler and Sarah Newbury focus on the contribution of the model organism Drosophila melanogaster to the understanding of RNA turnover. Since the factors controlling RNA decay are very similar in all animals the understanding of their function in the fruit fly Drosophila is relevant to that in other organisms, including humans. A particular advantage of this model organism is that the functions of ribonucleases can be assessed in the context of natural cells within tissues in addition to individual immortalized cells in culture.
In this review, the authors summarize the large body of Drosophila RNA stability research which demonstrates that the cytoplasmic decay machines, including the exosome, Dis3L2 and Xrn1, can regulate specific processes including apoptosis, proliferation, wound healing, and fertility. This is because some mRNAs important in these pathways appear to be particularly sensitive to specific decay pathways due to being “tagged” for destruction.
Drosophila research has also contributed to our understanding of how specific RNAs are targeted (or “tagged”) to particular degradation pathways. These targeting mechanisms include AU rich elements, miRNA targeting, and 3’ tailing. Elucidating the roles of cytoplasmic ribonucleases in degradation of particular transcripts is not only important for understanding Drosophila cellular pathways but is also relevant to human diseases such as cancer.
Kindly contributed by the Authors.

















