Globalization has increased health risks and created social alarms when dealing with readily transmissible infections. Among ‘global’ diseases, those caused by bacteria pose major threats to the population because of their high rates of morbidity and mortality, economic impact, and role as reservoirs of antibiotic-resistance (ABR) and virulence genes that are spread among them. Huge amounts of financial resources are devoted to treating community-acquired infections, like those acquired in hospitals (nosocomial infections), because of the high ABR levels found in the bacteria causing them (‘superbugs,’ in colloquial terms). Thus, it is extremely important to find and to validate common targets among ABR-bacteria that share global virulence regulatory genes and also to discover the ways for horizontal spread of ABR traits: knowledge of these matters will help to develop strategies to tackle dangerous infections.
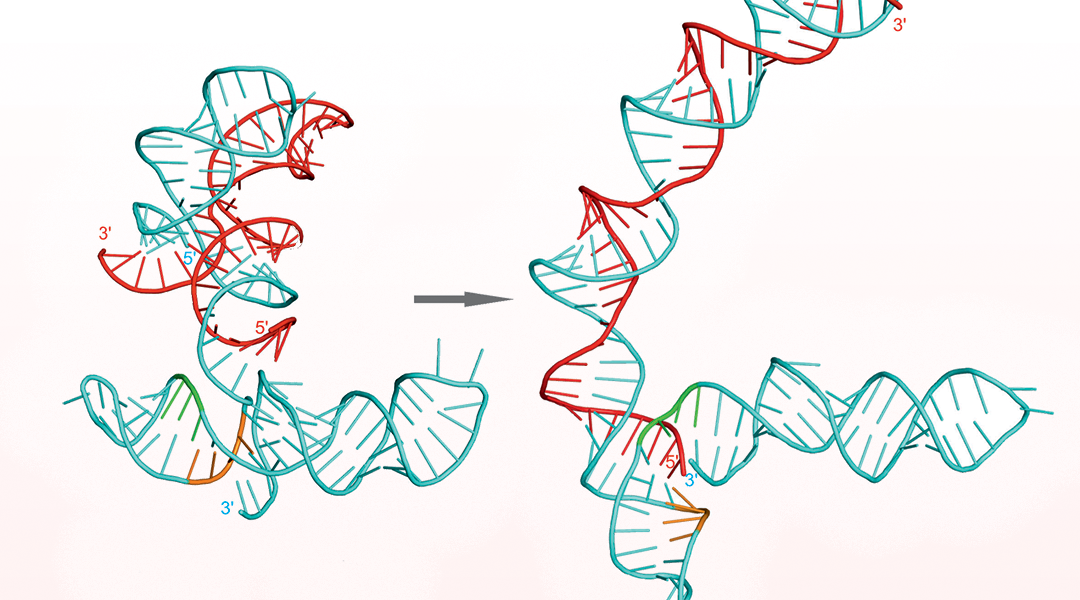
The spread of ABR traits is mediated by mobile genetic elements, especially those called plasmids, which are shared among bacteria and constitute a reservoir of common genes. Plasmids are DNA molecules able to multiply themselves (through replication or vertical transfer) independently of their bacterial host. Plasmids may also carry genes that allow them to colonize new hosts by functions specialized for their mobilization (conjugation or horizontal transfer). An important feature of plasmids is that their vertical transmission is tightly controlled by functions encoded by the plasmid itself, so that they keep a fixed number of copies within a given bacterial host. To achieve this control, plasmids may use different strategies, one of the most common being the employment of small antisense (as) RNAs.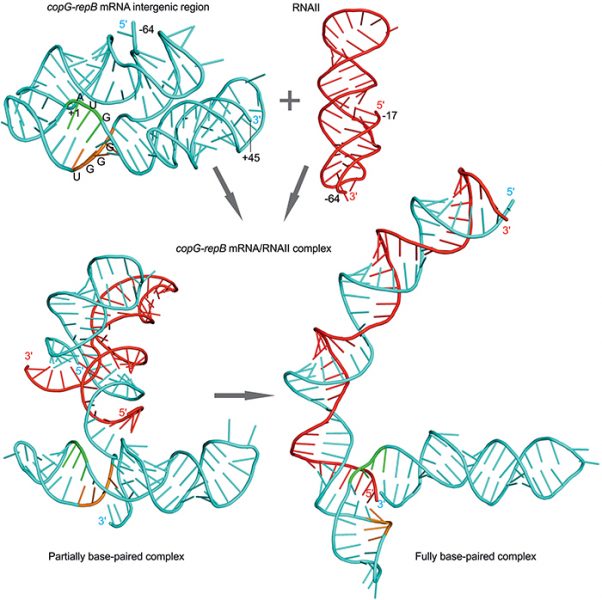
In a recent review in WIREs RNA, Pluta and Espinosa update previous and recent studies on the control of the number of copies exerted by rolling circle-replicating (RCR) plasmids by structured small antisense RNAs. They extend further their analysis by constructing novel structural models of these small antisense RNA molecules. RCR-plasmids constitute the smallest and most widespread members of the plasmid families. These tiny DNA packages lack the systems that allow them to segregate, that is the means by which their vertical transmission to daughter cells is secured.
How then these plasmids ensure their stable inheritance? They do so by producing many copies of themselves, assuring a happy end to their quest to perpetuate. Due to their small size, RCR-plasmids can afford high copy number while still not interfering with cell fitness. Nonetheless, they need to have efficient and (due to size constraints) compact machinery for keeping their genetic load to reasonable levels. RCR-plasmids achieve that goal by means of a small short-lived antisense RNA, alone or in combination with a plasmid-encoded transcriptional repressor protein. Be that as it may, small structured antisense RNAs play a critical role in the perpetuation of these important plasmid molecules, posing imaginative solutions to the ways the antisense RNAs can participate actively in the lifestyle of the bacterial plasmids.
Kindly contributed by the Authors.