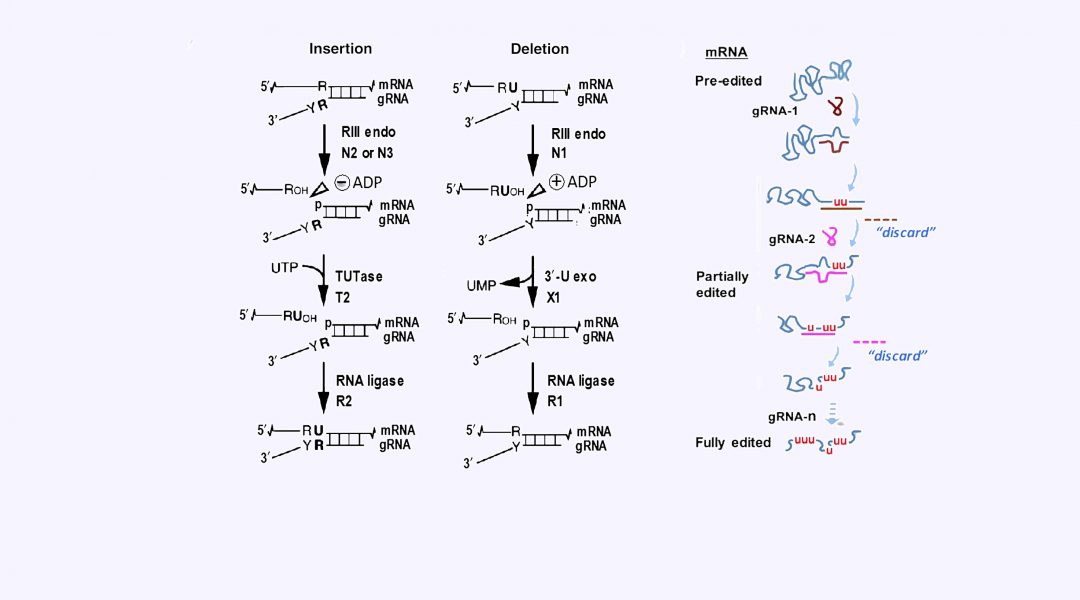
RNA editing causes massive remodeling of the mitochondrial mRNA transcriptome in trypanosomes and related kinetoplastid protozoa. This type of editing involves the specific insertion or deletion of uridylates (U) directed by small noncoding guide RNAs (gRNAs). Because U‐insertion exceeds U‐deletion by a factor of 10, editing increases the nascent mRNA size by up to 55%.
Trypanosomes and related kinetoplastid protozoa are early-branching eukaryotes with unique mechanisms of gene expression in both their nucleus and single mitochondrion. Massive RNA editing creates functional mitochondrial transcripts in these organisms. This editing involves specific uridylate (U) indels in a process of byzantine complexity that is developmentally controlled. In T. brucei, the U-indels are directed by ~40 proteins and a plethora of small non-coding guide RNAs (gRNAs). These components are organized in RNA holo-editosomes that assemble around mRNA and include macromolecular factors: the “RNA free” core editing enzyme, termed RECC complex, and several auxiliary RNPs including the RESC and REH2C complexes.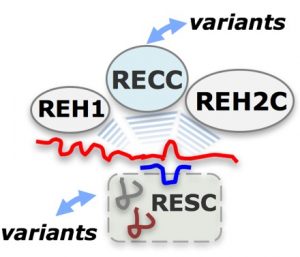
A review in WIREs RNA by Cruz-Reyes and colleagues discusses possible steps in the control of RNA editing which remains poorly understood. Past work suggests that it occurs during substrate recognition, the initiation and progression of editing, and during the life-cycle in different hosts. Recently discovered variants of the canonical macromolecular mRNA trans factors further complicates the model of RNA editing. This increased complexity also offers exciting opportunities for regulation and fine tuning in editing. In this review, authors examine specific examples of complex variants, differential effects of editing proteins on the mRNAs within and between T. brucei life stages, and possible control points in the holo-editosomes.
Kindly contributed by the Authors.