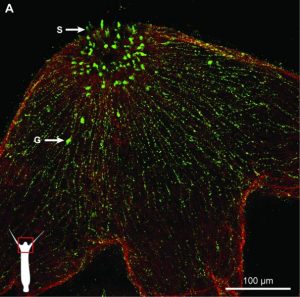
Hydra nervous system. RFamide-positive neurons (green) and muscular fibres of the epithelial cells as detected by phalloidin (red). G: ganglion neurons and S: sensory neurons.
The freshwater polyp Hydra possesses an anatomically simple nervous system. It is made up of about 6,000 nerve cells – both sensory and ganglion neurons – that are spread throughout the body without any signs of centralization. Recent studies have revealed an unexpected role of these nerve cells in shaping Hydra’s microbiome. In their review published in BioEssays, Alexander Klimovich and Thomas Bosch take a closer look at the intricate interplay between the Hydra polyp and its microbiota while particularly focusing on the role of the nervous system.
The interaction between Hydra and its microbiome is bi-directional, and this interaction influences diverse processes such as polyp contraction and feeding response. On the one hand, for example, germ-free Hydra polyps display strongly reduced and less spontaneous body contraction frequencies. This suggests that bacterial products may target the pacemaker neurons, which are responsible for the timing of contractions. On the other hand, neurons produce and secrete antimicrobial peptides that contribute to shaping the species-specific microbiome.
Investigating the microbiota-gut-brain axis in vertebrates has become rather popular recently. The presence of this microbiota-neuron interplay in Hydra may therefore offer a unique possibility to study this complicated interaction in a simple animal model organism to get a better understanding of the molecular mechanisms. In this regard, the authors also discuss some questions that need to be addressed to get further insights into mechanisms and function. One question is the exact nature of the microbial molecules that are recognized by Hydra’s neurons.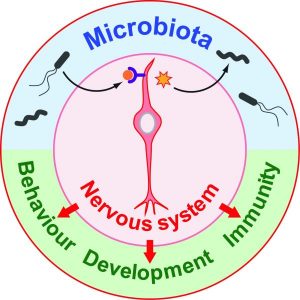
The authors propose that a systems biology approach with deep transcriptomic and metabolic analysis of the microbes associated with Hydra may help to identify the bacteria-derived neuroactive molecules. Furthermore, detailed knowledge of the neuronal receptors and signaling cascades is still lacking as well. To answer this question, a deep analysis of the transcriptome from Hydra nerve cells should be employed. In addition, the neuron-derived peptides and how they shape the microbiota also deserves further study. Answering these molecular and mechanistic questions will hopefully ultimately also help to get further information on the more general question of how the microbiota affects development.