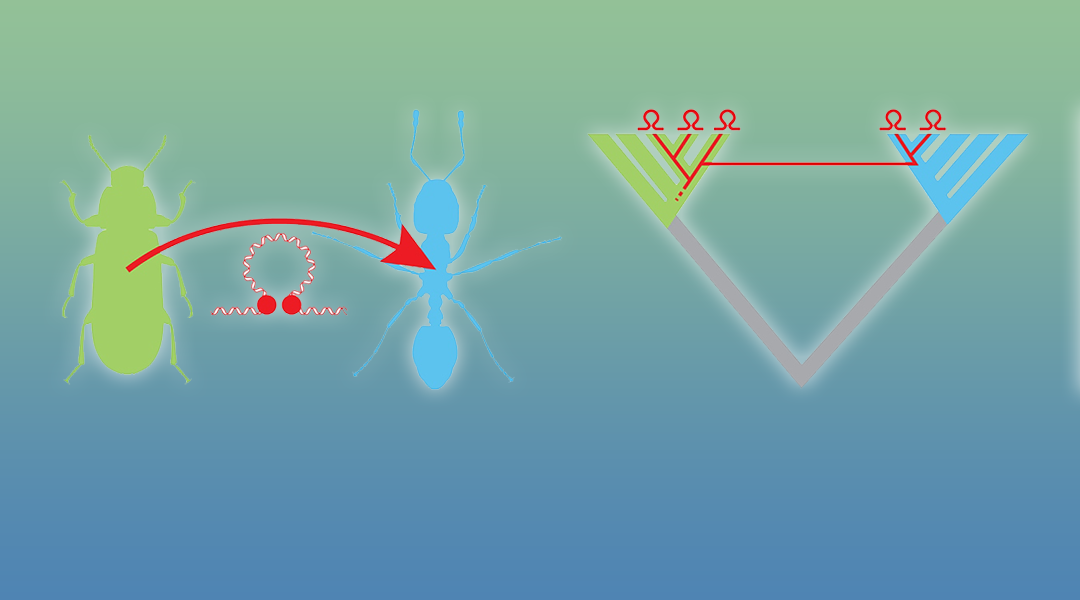
In contrast to “vertical” transfer where genetic material is transmitted from parent to offspring, horizontal transfer does not involve reproduction and may therefore also take place between organisms belonging to different species. This transfer may not only comprise genes (horizontal gene transfer or HGT), but may also include the transfer of transposable elements (horizontal transfer of transposable elements or HTT). By their nature, transposable elements can change their location in the genome, and HTT therefore plays an important role in genome evolution.
Despite this importance of HTT only few studies have so far analyzed this phenomenon at a large scale. In their review published in BioEssays, Jean Peccoud, Richard Cordaux and Clément Gilbert review and discuss the challenges that need to be overcome when one wants to undertake such an analysis.
In order to analyze the horizontal transfer of transposable elements (TEs), one first of all has to be able to reliably detect TEs in the genomes of a variety of different species. Accurately annotating TEs still requires manual verification steps and is therefore not easily feasible if one wants to compare many different genomes. In addition, the ability to detect TEs in the first place largely depends on the quality of the “source material”; i.e. the genomes being analyzed should be as complete as possible. In addition, fragmentation of the genome data will negatively impact the analysis as well. Another factor impacting the quality of the material is possible contamination by DNA from other organisms.
The next challenge arises around the question what TE sequences should be retained and compared. An analysis of HTT generally involves comparing consensus sequences of TEs. TEs are grouped together in a TE “family” if they show a certain degree of DNA sequence similarity over a specified portion of their length (both set at 80%). It is subsequently assumed that TEs from the same family resulted from one HTT event. However, the 80% grouping criterion is somewhat arbitrary and, additionally, it cannot be ruled out that a particular TE family is the result of multiple separate acquisition events.
Other challenges consist of how to reliably infer horizontal transfer and how to delineate transfer events. Here, the authors not only review the criteria that are being applied and the challenges associated with these criteria, but they also discuss possible ways of how to address these challenges.