Hfq is a ubiquitous RNA binding protein in bacteria that stabilizes small regulatory RNAs (sRNAs) and helps them bind with their mRNA targets. Hfq and sRNAs regulate how bacteria respond to stress or changes in their environment and are involved in the virulence of pathogenic bacteria such as Neisseria meningitidis, Vibrio cholerae, Borrelia burgdorferi and Listeria monocytogenes. Experiments from structural biology, biochemistry, and genetics have combined to reveal how Hfq plays matchmaker for sRNAs and mRNAs.
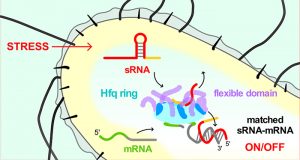
The chaperone Hfq speeds up match-making between small regulatory RNA (red) and mRNA (green/gray) in bacteria, helping bacteria respond to stress or changed growth conditions. The Hfq core (cyan) binds two RNAs, allowing complementary RNAs to base pair. Flexible regions of the Hfq protein (light blue) sweep RNA from the core, quickly cycling through prospective sRNA-mRNA pairs.
A bacterium may produce tens or hundreds of sRNA types that must act on their target mRNAs within a few minutes. Hfq speeds up the rate by which prospective sRNAs and mRNAs pair with each other in the test tube, and helps ensure that only the correct sRNA-mRNA pairs are formed in the cell. This rapid response allows the bacterium to react to different environmental signals in a timely manner. An mRNA may be regulated by more than one sRNA, with each sRNA being induced by a different trigger. Additionally, sRNAs compete against each other for a limited quantity of Hfq protein, so that a high level of one sRNA may interfere with the activity of another sRNA. These characteristics allow multiple stress signals to control the output of a target gene via NOT, OR and NAND logic gates.
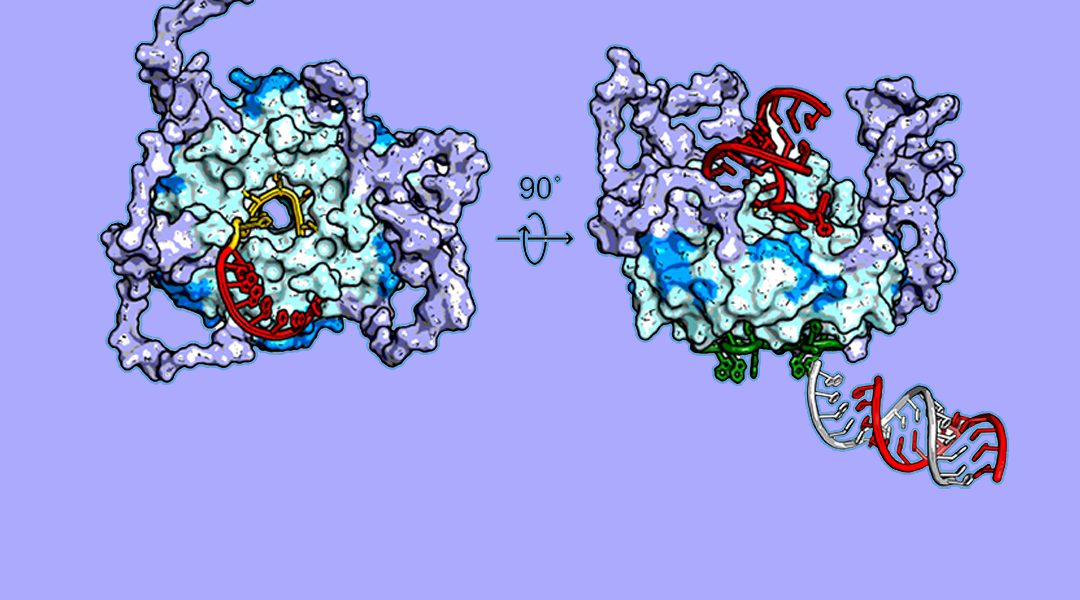
The ring-shaped Hfq protein can bind one RNA on each face. Positively charged arginine residues on the outer edge of the ring promote base pairing between complementary RNAs. Remarkably, flexible “tails” extending from the edge of the ring sweep RNA away, prevent mismatched RNA pairs from remaining bound to the Hfq ring and recycle Hfq to act on other sRNAs. The interplay between structured and flexible domains of Hfq optimizes sRNA regulation, and is common among RNA chaperones. These flexible domains appear to function as independent modules, enabling predictions for how Hfq homologs may act in different species of bacteria.
Read more in a review article recently published in WIREs RNA.
Kindly contributed by the Authors.