Life on Earth appears in thousands of shapes and forms, from simple single-celled bacteria and yeasts to complex animal bodies in which the function of each organ is carefully orchestrated, from the brown fruiting bodies of certain slime molds to the colorful patterns of the peacock’s tail. Some animals live solitary lives, caring only about their own direct descendants, or laying eggs in the ground and leaving them to their destiny, while others live in groups, like honeybees, where the workers feed the queen’s offspring and defend the nest against intruders. But when researchers study biological organisms on the molecular level, all life appears similar: genetic information is stored in DNA, and it is transferred into a functional end product, usually a protein, through a sequence of processes, starting with the transcription of DNA into messenger RNA. How then, do the striking differences between various life forms arise? What makes a multicellular mushroom different from unicellular yeast? What makes social honeybees different from their solitary relatives, and what distinguishes a human from a chimpanzee? This question has long puzzled biologists.
One answer may lie in the differences of how this information transfer from DNA to protein is regulated. All cells in the human body, for example, contain the same genetic information. But the same genetic information can produce different cell types, organs and structures, by regulating which parts are expressed and the expressed proteins are produced in smaller or larger quantities. Even single genes in higher organisms are not fixed blueprints for specific proteins, they are more like recipes composed of different parts which can be combined flexibly. The same recipe can be used to produce a double cheeseburger, a low-carb burger wrapped in lettuce or a vegan tofu burger, depending on the demands of the customer, or, in the case of the cell, the demands of the organism or the physiological and developmental signals received from the environment. It is also possible that the differences between species can often be found in the way these recipes are read and executed rather than the components of the recipes themselves. In other words, gene regulatory differences might often be more important than differences in protein sequence and structure.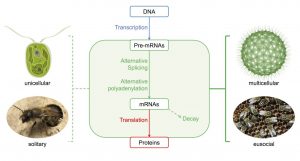
In a new review article in WIREs RNA, Schaefke et al. zoom in on the steps occurring after the transcription of DNA into RNA, so called post-transcriptional regulation, and how these steps have evolved to cause differences between species. First, non-coding pieces, the introns, are cut out of the messenger RNA, and the protein-coding parts plus regulatory sequences on both ends are joined together. This process is called splicing, and sometimes can lead to different mature messenger RNAs, depending on exactly which parts are combined. Therefore, alternative splicing ultimately can lead to different protein end products. Next, a tail of multiple adenines (the ‘A’ in the genetic 4-letter-code) is attached to the end of the messenger RNA, and this addition can happen at different locations, which can in some cases also lead to different protein products, and in other cases affect the stability or localization of the RNA molecule. Finally, the mature messenger RNA is translated into protein. The quantity of each protein depends on translation efficiency and on the amount of RNA available as a template for protein production. This in turn depends not only on the transcription rate, but also on how fast specific messenger RNA molecules are degraded, a highly regulated process. By increasing the potential of the cell to choose different developmental pathways even with limited genetic material, all of these post-transcriptional processes, splicing, polyadenylation, translation and decay, might have contributed to the evolution of complex life forms, including the formation of multicellular organisms or social groups from smaller units.