In 2004, Edward Boyden and Karl Deisseroth made a groundbreaking discovery. The US scientists had just placed neurons cultured in a petri dish under a conventional microscope. However, these cells were not just regular neurons; they expressed channelrhodopsin-2, a light-gated cation transporter naturally found in flagellate algae. Boyden started to shine patterns of brief blue light pulses onto the genetically modified neurons. Remarkably, the cells swiftly responded by firing action potentials precisely mimicking the pattern of the light stimulus. Optogenetics was born and soon revolutionized our abilities to study how behavior, memory, and feelings emerge from the complex interplay of nerve cells in our brains.
It quickly became apparent that the concept of controlling the activity of genetically encoded proteins in living cells with light would not only revolutionize neurosciences research. Driven by the rapid discovery and characterization of light sensing proteins derived from plants, fungi, and bacteria, optogenetics soon expanded toward diverse cell biology areas. Today, this technology is employed for studying all different kinds of dynamic cellular signals governing, for instance, motility, stress responses, development, differentiation and death in various cell types and species.
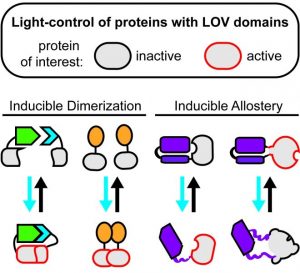
One of the major drivers in the rapidly progressing, non-neuronal optogenetic field are LOV (for light-oxygen-voltage) domains. This highly diverse class of small blue light sensors can be found, e.g., in phototropins, the proteins mediating the plant’s movement towards the sun (phototropism). By artificially coupling LOV domains to cellular proteins or signaling peptides, customized optogenetic input modules can be created that facilitate precise perturbations of selected cellular pathways and functions.
In a review article published in Advanced Biosystems, Hoffmann et al. discuss the broad range of applications enabled by LOV domains. The authors describe the great portfolio of strategies available to confer optogenetic regulation on selected signaling factors, transcriptional regulators, receptors, or enzymes via LOV-mediated, light-dependent protein association, dissociation, allostery, or recruitment to selected subcellular loci. The article provides a comprehensive introduction into the utility of LOV domains to newcomers in the non-neuronal optogenetics and aids researchers in the selection of LOV-based optogenetic tools suitable for specific experimental settings and scientific questions.
Kindly contributed by the authors.