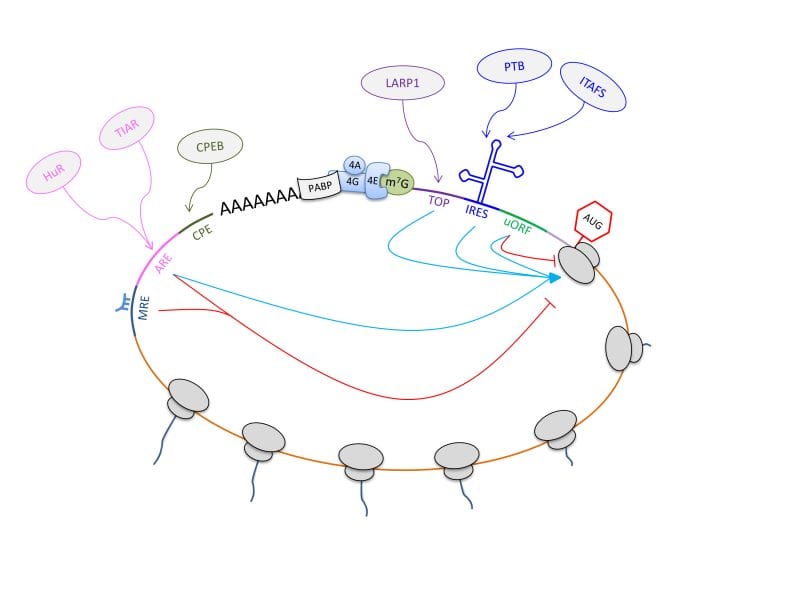
From the moment it is synthesized in the nucleus, each RNA molecule is found in association with a range of RNA-binding proteins (RBPs), the combination of which will determine the fate of the transcript. Translation, the synthesis of proteins from a template messenger RNA performed by ribosomes, depends heavily on RBPs for its thorough modulation, which is of fundamental importance for cell homeostasis as well as the response to external cues.
In a recent Open Access advanced review in WIREs RNA, “Trans-acting translational regulatory RNA binding proteins,” Harvey and colleagues integrate the fundamentals of RBP-mediated translation regulation with some of the latest, most exciting discoveries.
Tremendous progress has been made in recent years, with newly developed methodologies bringing to light an outstanding amount of information about RBPs and the RNAs they bind. Throughout the review, the most relevant among these techniques are discussed in detail, with their advantages and shortcomings, and some missing tools are identified that could prove key to the advancement of current knowledge.
Four important RBPs – LARP1, PTB, CPEB, and PABP – are described thoroughly and often used as a starting point to discuss canonical and noncanonical RNA-binding domains, as well as relevant RNA motifs. Particular attention is also given to aspects that have only recently emerged as important players in translational regulation. For example, the authors review recent reports suggesting that ribosomal subunit heterogeneity might confer RNA specificity and that post-translational modifications might modulate the activity of RBPs and ribosomal proteins.
Kindly Contributed by the Authors.