As a result of earth’s rotation on its axis, living organisms are confronted by daily changes of their environment that follow the day and night cycle. To be able to anticipate those changes by developing an internal clock represents an evolutionarily conserved adaptation that can be traced back to the earliest life forms. In mammals, this endogenous clock controls most biological functions, which then oscillate with a period close to 24h, which is called circadian. These circadian rhythms that take place from cellular to physiological and behavioral levels are mostly driven by a rhythmic gene expression. In each tissue, 8-40% of expressed genes follow a circadian expression pattern.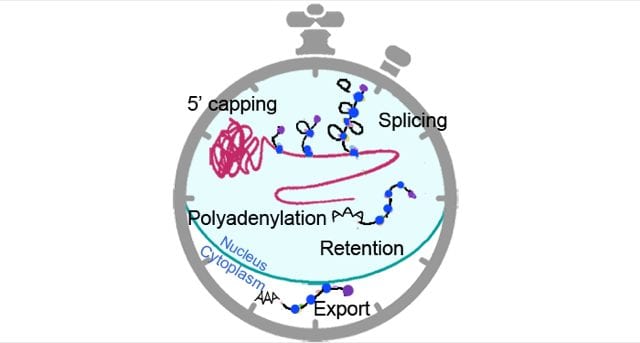
While circadian oscillators are based on a molecular mechanism that involves transcriptional-translational feedback loops that control the rhythmic transcription of many clock-controlled genes, recent studies have unexpectedly shown that 80% of rhythmic genes are clock-controlled by mechanisms occurring after transcription.
Many steps occur between transcriptional initiation and protein function and many, if not all, of these steps of the RNA life cycle are subject to circadian regulation. These steps include all processes involved in RNA maturation like capping, splicing and 3’ end termination. RNA transport can also follow a circadian pattern, with a circadian nuclear retention driven by rhythmic expression within the nucleus of particular bodies (the paraspeckles) and circadian export to the cytoplasm driven by rhythmic proteins acting like cargo.
Finally, RNA degradation may also follow a circadian pattern through the rhythmic involvement of miRNAs. In a recent review in WIREs RNA, “Circadian processes in the RNA life cycle,” the authors summarize the current knowledge of the post-transcriptional circadian mechanisms known to play a prominent role in shaping circadian gene expression in mammals.
Kindly contributed by the Authors.
















