How stem cells differentiate into the myriad diverse cell types that ultimately form tissues has been an area of extensive research. Studying this process at the population level unfortunately masks rare or transient intermediates. The application of ever more sophisticated single cell technologies, however, is able to address this problem.
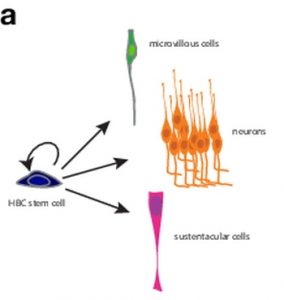
In their article published in BioEssays, Russell Fletcher, Diya Das and John Ngai particularly discuss one such single cell technology, namely single-cell RNA-sequencing. They review how an integration of this technology with clonal lineage tracing provides a robust strategy for studying how an individual stem cell changes through time to differentiate and self-renew. The authors exemplify this by taking a look at the olfactory epithelium where horizontal basal cell (HBC) stem cells differentiate into microvillous cells, neurons and sustentacular cells.
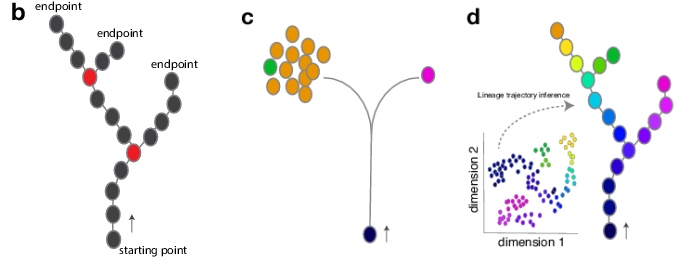
In general, a multipotent stem cell can give rise to multiple cell fate endpoints as shown in panel b below. In classic genetic lineage tracing, a stem cell is labeled, and the labeling is detected in its descendants. This provides information about the stem cell fate potential; i.e. what type of differentiated cell this labeled stem cell might become (see panel c). However, lineage tracing does not allow the identification of intermediate stages along a lineage trajectory nor is it able to pinpoint branch points. Single-cell RNA-sequencing (scRNA-seq), on the other hand, can discriminate the different cell types present in a heterogenous population. As shown in panel d, lineage trajectory inference tools can then be used on data from a scRNA-seq experiment (visualized in a reduced dimension plot) to predict stem cell lineage trajectories as well as branch points.
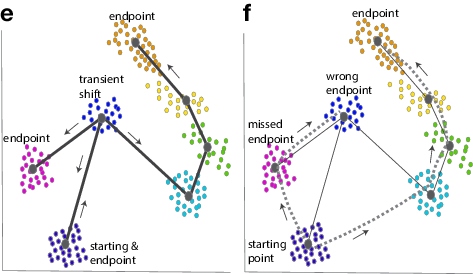
Lineage trajectory inference tools rely on the assumption that cells that are more similar in gene expression are closer together on a lineage trajectory. In addition, these tools also make the assumption that the paths are unidirectional. Both of these assumptions may create problems as exemplified in panel f below where the dashed lines indicate a wrong order in the lineage trajectory. Therefore, any lineage trajectory models obtained from scRNA-seq data need to be validated experimentally. Nevertheless, an integration of these two approaches – lineage tracing and scRNA-seq – provides a robust strategy to gain further insights into the cell fate changes and the paths along which stems cells differentiate.