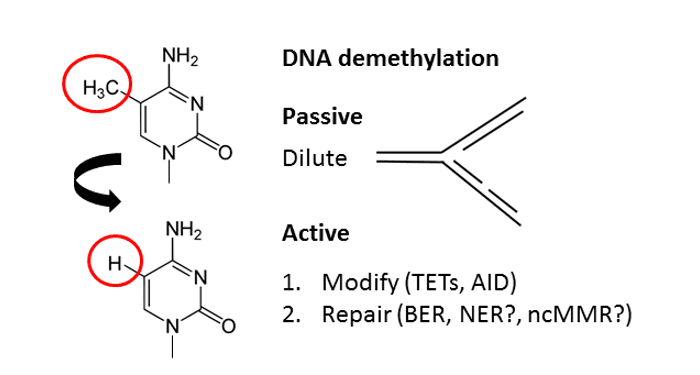
In their review in BioEssays, Matthias Bochtler et al. discuss the current state of knowledge surrounding active DNA demethylation pathways.
In general, DNA demethylation can occur either passively or actively. The passive process takes place during DNA replication when the newly synthesized strand is not methylated and the methylation mark is thus “diluted” over time. Active DNA demethylation is independent of DNA replication and requires the action of enzymes. Despite earlier claims, direct demethylation of 5-methylcytosine (5mC) to cytosine does not occur. Instead, all known in vivo conversions of 5mC to cytosine involve replacement of the methylated nucleotide.
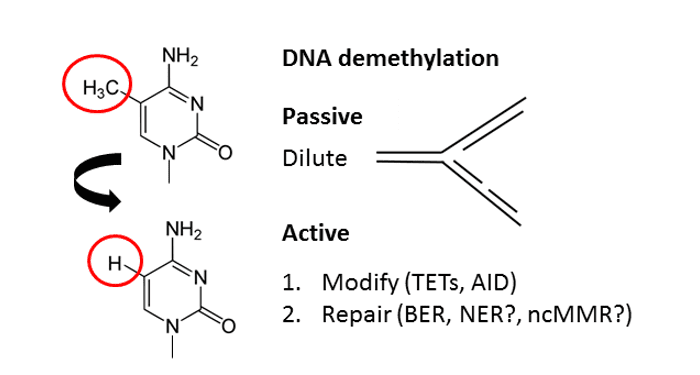
The first step in this process is DNA base modification: either TET-mediated oxidation of the methylated base or deamination of the methylated (or a nearby base) by AID (activation induced deaminase). TET (ten-eleven-translocation) proteins not only convert 5mC to 5-hydroxymethylcytosine (5hmC), but also 5hmC to 5-formylcytosine (5fC), and 5fC to 5-carboxylcytosine (5caC). AID, on the other hand, deaminates cytosine to uracil and – to a lesser extent – 5mC to thymine. In that way, AID could either act directly on 5mC or indirectly by modifying neighboring, ‘regular’ cytosines.
The second step of active DNA demethylation is nucleotide replacement: The modified nucleotide (possibly together with surrounding nucleotides) is replaced via DNA damage repair pathways, presumably mainly by base excision repair. Various glycosylases, such as thymine DNA glycosylase (TDG) are involved in this process. Despite recent progress in deciphering these active DNA demethylation pathways, quite a few interesting questions remain; for example how loci are targeted for demethylation or when the replacement of nucleotides is initiated by TET and when by AID.