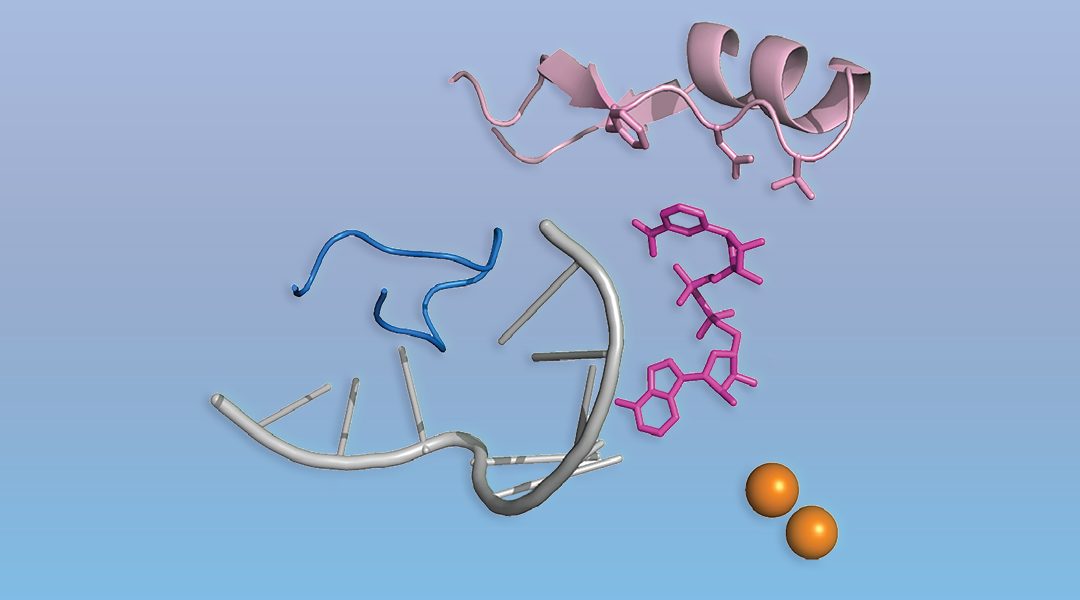
The process of gene expression starts with the production of an RNA copy of a gene. Only in eukaryotes (or so it was thought) does this RNA bear a cap, the special molecule m7G at the 5’ end. The cap protects RNA from degradation and directs it to the protein-making machinery. A set of dedicated enzymes then places the m7G cap onto RNA. This dogma is now shaken by a recent discovery of an universal non-canonical RNA capping, present in all kingdoms of life. In a WIREs RNA review, authors Christina Julius and Yulia Yuzenkova examine this recent discovery by describing non-canonical RNA capping along with molecular mechanisms of cap addition and removal. The authors further discuss a wide range of potential downstream functional consequences of non-canonical capping.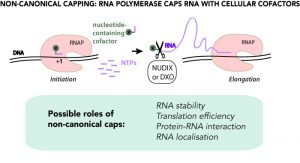
Despite superficial similarities of the new type of capping with canonical capping, there are striking differences. First, there is no particular capping molecule, and a variety of cellular cofactors participating in metabolism and redox processes, including NAD+/NADH, can serve as caps. Second, no dedicated machinery is needed. The main RNA making enzyme (RNA polymerase) caps RNA by itself using cofactors as the first building block of an RNA chain. Potentially any molecule containing nucleotide moiety can serve as non-canonical cap, including even cell wall precursors.
As of now, non-canonical capping has been found in bacteria, such as E.coli and B.subtilis, and in yeast and human cells, including their mitochondria. This very universality brings the question of whether non-canonical capping is a regulated or just a stochastic process, side reaction of transcription. In any case, being at the intersection of metabolism and core gene expression, non-canonical capping is bound to affect cell physiology. The physiological role of non-canonical capping is a subject of intense research. Non-canonical capping is already shown to affect the stability of RNA, by analogy with classic capping. Other proposed roles range from redox state-dependent regulation of gene expression to RNA localization.
Kindly contributed by the Authors.