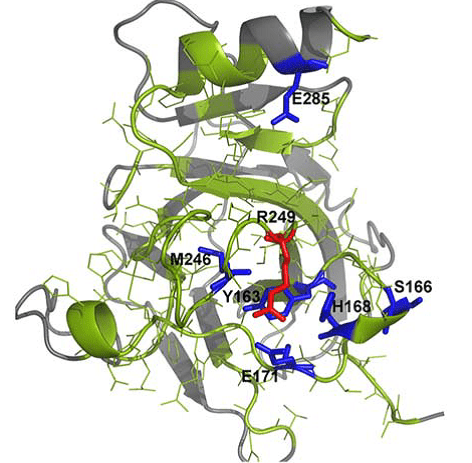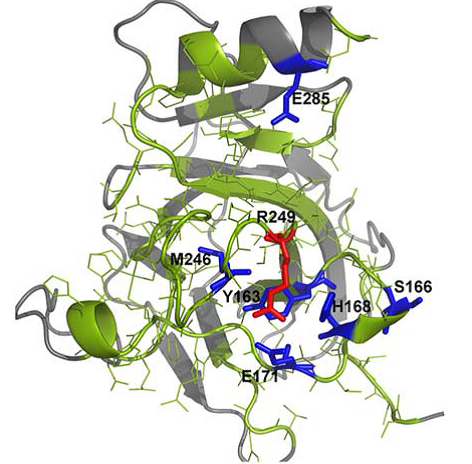
Spatial distribution of amino acid residues in tumor suppressor protein p53 after in silico mutation.
The use of bioinformatics tools in answering key questions in biology is becoming increasingly common. Thanks to their massive potential to hold and process data whether in bytes or base pairs. Electrostatic potential of proteins determines their ability to interact with specific ligands and other macromolecules. Hence it is a trait of biological significance.
Amino acids are building blocks of a protein. Mutation of amino acids in a protein significantly alters the electrostatic potential of proteins. Site directed mutagenesis is the process of introducing targeted mutations in the double stranded DNA. Often this technique involves inducing changes in parts (amino acid residues) of a protein, and then deciding the role of amino acids that underwent the change. If induced directly in biological systems, these changes could lead to undesirable outcomes. Therefore, researchers need an experimental environment in which they can conduct these tests, and then understand the outcome. It is at this juncture that in silico tools come in handy
In a recent issue of the Journal of Computational Chemistry, Valdebenito-Maturana and colleagues, discuss the use of a new in silico simulation platform Mutantelec for comparative electrostatic potential profiling of proteins. Using Mutantelect, the authors elucidate mutations in tumor suppressor protein p53 that would inhibit its ability to bind to a target DNA sequence.
Software that is currently used for this purpose does not consider the net effect on the electrostatic potential of target proteins, or the global charge distribution of mutant in silico variants of the protein of interest. As a result, minor changes that happen in single amino acid residues in a protein might go unnoticed. On the other hand, Mutantelect predicts and evaluates changes in charge distribution of a mutated protein, while also being freely available and user friendly. The sensitivity of Mutantelect to pick up critical details, enables understanding of even subtle changes such as modification of a single amino acid residue in a protein structure. Functions such as these can be extremely useful in understanding mutations in enzymes that could cause metabolic diseases.