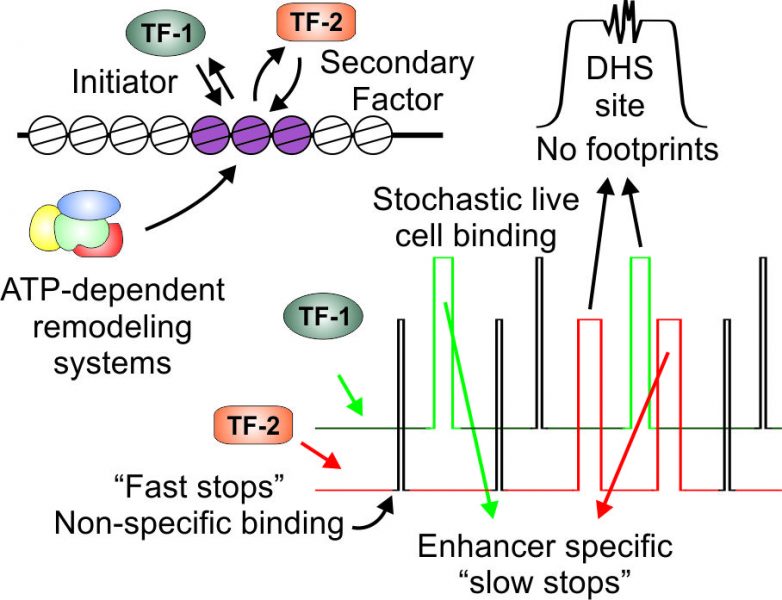
The identity of each cell type in the body and the response of cells to a constantly-changing environment are largely defined by controlling which genes are expressed in a specific cell type at a given time point. The regulation of gene expression is dictated by transcription factors (TFs) that bind specific elements in genomic DNA termed enhancers. Advances in genome-wide technologies have enabled the enumeration and characterization of the enhancer repertoire bound by TFs. These findings are based on averaging the signal from millions of cells and have led to a model whereby 1) TFs are bound on DNA for minutes to hours, and 2) complex structures composed of many TFs are simultaneously bound at the enhancer elements. In a recent review article in WIREs Systems Biology and Medicine, Goldstein and Hager describe two technological breakthroughs that undermine this static model of TF binding to enhancers.
Traditional techniques rely on assessing a population of cells that was ‘frozen’ in time (this is achieved by a chemical covalently crosslinking TF-DNA contacts). In contrast, fluorescent microscopy techniques image TF behavior in live cells and along a time axis. These techniques reveal a highly dynamic binding behavior for TFs. In particular, single molecule tracking has recently evolved as a promising methodology to probe enhancer function in live cells by tracking individual TF molecules in real time. Collectively, data from various live cell imaging approaches refuted the assumption that TFs reside on DNA for long periods of time. Rather, TFs dynamically exchange with their binding sites on the time scale of seconds. These findings are complemented by advances in ‘TF footprinting’. TFs that bind specific enhancers sometimes leave a ‘footprint’ on the bound region by protecting the DNA from enzymatic cleavage. Genome-wide footprinting efforts demonstrate that while some TFs leave footprints when bound to DNA, others do not. These footprint-lacking TFs are nonetheless bound at the relevant enhancers. The dynamic nature of TF binding is a plausible explanation for footprint-lacking TFs. Thus, when a factor dynamically exchanges on and off DNA, it cannot efficiently protect it from cleavage. Indeed, the authors describe the correlation between the DNA residence time of TFs and their footprinting capacity.
In their review article, Goldstein and Hager discuss the implications of these new approaches for an accurate understanding of enhancer function in real time. Integration of findings from the many approaches now employed in the study of enhancer function suggest a highly dynamic view for the action of enhancer activating factors.
Text contributed by the authors.