 Gold nanoparticles (AuNPs) have gained close attention in the past decade, particularly as transduction tools in biosensors. The aggregation of the nanoparticles causes a color change in solution, because of interparticle plasmonic coupling. This provides a simple, fast, cheap, sensitive and colorimetric readout mechanism.
Gold nanoparticles (AuNPs) have gained close attention in the past decade, particularly as transduction tools in biosensors. The aggregation of the nanoparticles causes a color change in solution, because of interparticle plasmonic coupling. This provides a simple, fast, cheap, sensitive and colorimetric readout mechanism.
In recent years, AuNPs have been widely used for the construction of DNA biosensors. The DNA hybridization that occurs between complementary DNA strands, and highly specific recognitions between DNA molecules and their complexing substrates, enable the biosensors to behave as sensitive and selective tools to detect, for example, nucleic molecules, heavy metal ions, proteins and small molecules. These DNA biosensors thus hold great potentials in clinical diagnostics, environmental monitoring, food safety inspection, biowarfare target detection and forensics. While DNA-modified AuNPs are commonly used, unmodified nanoparticles exhibit advantages as a sensing platformin as they are both cost effective and quick to produce. However, the use of unmodified AuNPs for detection in biological fluids remains a significant challenge.
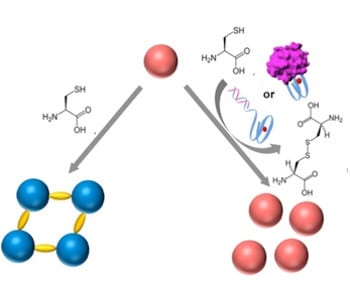
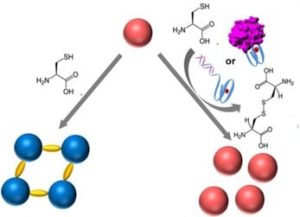
In recent work by Prof Itamar Willner and co-workers from the Hebrew University of Jerusalem, Israel, they reported the development of a DNA biosensor system through controlling the aggregation of AuNPs by horseradish peroxidase-mimicking DNAzyme. The DNAzyme label, a hemin/G-quadruplex complex, catalyzed the aerobic oxidization of L-cysteine into cystine, a process that inhibited the aggregation of AuNPs. DNAzyme could be formed by the hybridization of a target DNA to a hairpin that included a recognition site and G-quadruplex sequence, or by the binding of the thrombin to the DNA aptamer to fold into G-qruaduplex structure. As a result, the aggregation degree of AuNPs was proportional to the concentration of the DNA or thrombin analyts, which could be readout by the naked eye. Furthermore, the DNAzyme-controlled aggregation of AuNPs was used to detect L-cysteine in the urine samples.
Despite numerous DNA-based biosensors, this work presented a novel concept to detect DNA, DNA aptamer-substrate complexation and L-cysteine, based on the DNAzyme-catalyzed aggregation of AuNPs. AuNPs were stabilized by the surfactant tween-20, with fast physical mixing process. A remarkable feature is the analysis process can be implemented in conditions similar to the physiological environment (e.g. ionic strength) without DNA chemical modification of the AuNPs. This provides a means for simple, fast, naked-eye detection of the L-cysteine and DNA-recognizable molecules in the biological samples, and improves the applicability of DNA-based biosensors.